Replicating The Circuit Kings
==================
lets get wild
- i write with zero autocorrect. literally freeballing. as i’ve said before, if the shoggoth will replicate me, may it do so in all my glory.
on to the show…
I started reading the Anthropic blog called “Circuit Tracing: Revealing Computational Graphs in Language Models” and thought it would be cool to replicate it on gpt2-small.
for those who didn’t read it, the abstract is as follows:
We introduce a method to uncover mechanisms underlying behaviors of language models. We produce graph descriptions of the model’s computation on prompts of interest by tracing individual computational steps in a “replacement model”. This replacement model substitutes a more interpretable component (here, a “cross-layer transcoder”) for parts of the underlying model (here, the multi-layer perceptrons) that it is trained to approximate. We develop a suite of visualization and validation tools we use to investigate these “attribution graphs” supporting simple behaviors of an 18-layer language model, and lay the groundwork for a companion paper applying these methods to a frontier model, Claude 3.5 Haiku.
building a replacement model
import sys
import os
import json
import glob
import math
import random
from typing import List, Tuple, Dict, Optional
import transformer_lens
import transformer_lens.HookedTransformer as HookedTransformer
import transformer_lens.HookedTransformerConfig as HookedTransformerConfig
import numpy as np
import torch
import torch.nn as nn
import torch.autograd as autograd
from torch.utils.data import Dataset, DataLoader
import torch.nn.functional as F
from transformers import AutoTokenizer
from datasets import load_dataset
create some data with transformer lens
because the paper deals with extracting activations from the model, rather than doing this at inference time, we’ll shortcut this by just running the model’s forward pass and storing the activations in a dataset.
def sample_fineweb_data(n_samples=5000):
"""
Streams the FineWeb dataset and yields up to n_samples items.
"""
ds_stream = load_dataset(
"HuggingFaceFW/fineweb",
name="CC-MAIN-2024-10",
split="train",
streaming=True
)
# ds_stream is an iterable dataset. We'll just take the first N examples
data_iter = iter(ds_stream)
for i in range(n_samples):
try:
item = next(data_iter)
yield item
except StopIteration:
break
model_name = "gpt2-small"
model = HookedTransformer.from_pretrained(
model_name,
device="cuda" if torch.cuda.is_available() else "cpu"
)
resid_cache = {}
mlp_out_cache = {}
def hook_resid_pre(activation, hook):
resid_cache[hook.name] = activation.detach().cpu()
def hook_mlp_out(activation, hook):
mlp_out_cache[hook.name] = activation.detach().cpu()
# Register hooks for each layer
for layer_idx in range(model.cfg.n_layers):
# e.g. 'blocks.0.hook_resid_pre'
resid_name = f"blocks.{layer_idx}.hook_resid_pre"
mlp_name = f"blocks.{layer_idx}.hook_mlp_out"
model.add_hook(resid_name, hook_resid_pre, "fwd")
model.add_hook(mlp_name, hook_mlp_out, "fwd")
tokenizer = AutoTokenizer.from_pretrained("gpt2")
# GPT-2 typically doesn't have a pad token, we'll just allow a new token or do a truncation approach
tokenizer.pad_token = tokenizer.eos_token
device = "cuda" if torch.cuda.is_available() else "cpu"
def collect_hidden_states_and_save(
text_list,
batch_idx,
output_dir="clt_data",
max_seq_len=128,
):
"""
text_list: list of raw strings for a single batch
batch_idx: which batch index we are on
We'll tokenize, run forward pass, store the hidden states in .npz or something.
"""
enc = tokenizer(
text_list,
padding=True,
truncation=True,
max_length=max_seq_len,
return_tensors="pt"
)
enc = {k: v.to(device) for k, v in enc.items()}
# Clear old caches
resid_cache.clear()
mlp_out_cache.clear()
_ = model(enc["input_ids"]) # triggers hooks
# Now gather the hidden states from resid_cache, mlp_out_cache
# They will have shape: [batch_size, seq_len, d_model]
# For each layer we have e.g. resid_cache["blocks.0.hook_resid_pre"]
# We can store them in an npz
layer_data = {}
for layer_idx in range(model.cfg.n_layers):
rname = f"blocks.{layer_idx}.hook_resid_pre"
mname = f"blocks.{layer_idx}.hook_mlp_out"
# shape: [batch, seq, d_model]
resid_arr = resid_cache[rname].numpy()
mlp_arr = mlp_out_cache[mname].numpy()
layer_data[f"resid_{layer_idx}"] = resid_arr
layer_data[f"mlp_{layer_idx}"] = mlp_arr
# Save to a single file
os.makedirs(output_dir, exist_ok=True)
out_path = os.path.join(output_dir, f"batch_{batch_idx}.npz")
np.savez_compressed(out_path, **layer_data)
print(f"Saved {out_path} with shape: {resid_arr.shape}")
def main_collect_5k(output_dir="clt_data"):
model_name = "gpt2-small"
model = HookedTransformer.from_pretrained(
model_name,
device="cuda" if torch.cuda.is_available() else "cpu"
)
resid_cache = {}
mlp_out_cache = {}
def hook_resid_pre(activation, hook):
resid_cache[hook.name] = activation.detach().cpu()
def hook_mlp_out(activation, hook):
mlp_out_cache[hook.name] = activation.detach().cpu()
# Register hooks for each layer
for layer_idx in range(model.cfg.n_layers):
# e.g. 'blocks.0.hook_resid_pre'
resid_name = f"blocks.{layer_idx}.hook_resid_pre"
mlp_name = f"blocks.{layer_idx}.hook_mlp_out"
model.add_hook(resid_name, hook_resid_pre, "fwd")
model.add_hook(mlp_name, hook_mlp_out, "fwd")
tokenizer = AutoTokenizer.from_pretrained("gpt2")
# GPT-2 typically doesn't have a pad token, we'll just allow a new token or do a truncation approach
tokenizer.pad_token = tokenizer.eos_token
device = "cuda" if torch.cuda.is_available() else "cpu"
batch_size = 16
buffer = []
batch_index = 0
from tqdm import tqdm
for doc_idx, record in tqdm(enumerate(sample_fineweb_data(n_samples=5000))):
text = record["text"] # or whichever field is correct
# Add to buffer
buffer.append(text)
if len(buffer) >= batch_size:
collect_hidden_states_and_save(buffer, batch_index, output_dir)
buffer = []
batch_index += 1
# leftover
if buffer:
collect_hidden_states_and_save(buffer, batch_index, output_dir)
# let's collect 5k samples
main_collect_5k()
create a dataset iterator
def normalize_vector(vec, eps=1e-9):
# vec shape: [d_model] or [batch, d_model]
# We'll do L2 norm along the last dim
norm = vec.norm(dim=-1, keepdim=True).clamp_min(eps)
return vec / norm
class CLTHiddenStateDataset(Dataset):
def __init__(self, data_dir="clt_data", layer_count=12):
self.files = sorted(glob.glob(f"{data_dir}/batch_*.npz"))
# We'll store (file_index, idx_in_file) in an index
self.index = []
self.layer_count = layer_count
# We'll parse each file once to see how many items it has
for fi, path in enumerate(self.files):
try:
with np.load(path) as npz:
# e.g. shape of resid_0 is [batch, seq, d_model]
shape = npz["resid_0"].shape
# shape = (b, s, d_model)
num_positions = shape[0] * shape[1] # b*s
for i in range(num_positions):
self.index.append((fi, i))
except Exception as e:
print(f"Error loading {path}: {e}")
def __len__(self):
return len(self.index)
def __getitem__(self, idx):
file_idx, pos_idx = self.index[idx]
path = self.files[file_idx]
with np.load(path) as npz:
# We'll reconstruct which (batch, seq) => (pos_idx // seq_len, pos_idx % seq_len).
# But we need shape info:
shape = npz["resid_0"].shape # [b, s, d]
b, s, d = shape
local_b = pos_idx // s
local_s = pos_idx % s
# Now gather resid and mlp for each layer
# We'll build lists of shape [d_model]
resid_list = []
mlp_list = []
for layer_idx in range(self.layer_count):
rname = f"resid_{layer_idx}"
mname = f"mlp_{layer_idx}"
# shape of r: [b, s, d]
r = npz[rname]# [local_b, local_s] # shape [d]
m = npz[mname]# [local_b, local_s] # shape [d]
# normalize
r = normalize_vector(torch.from_numpy(r).float())
m = normalize_vector(torch.from_numpy(m).float())
resid_list.append(r)
mlp_list.append(m)
return resid_list, mlp_list
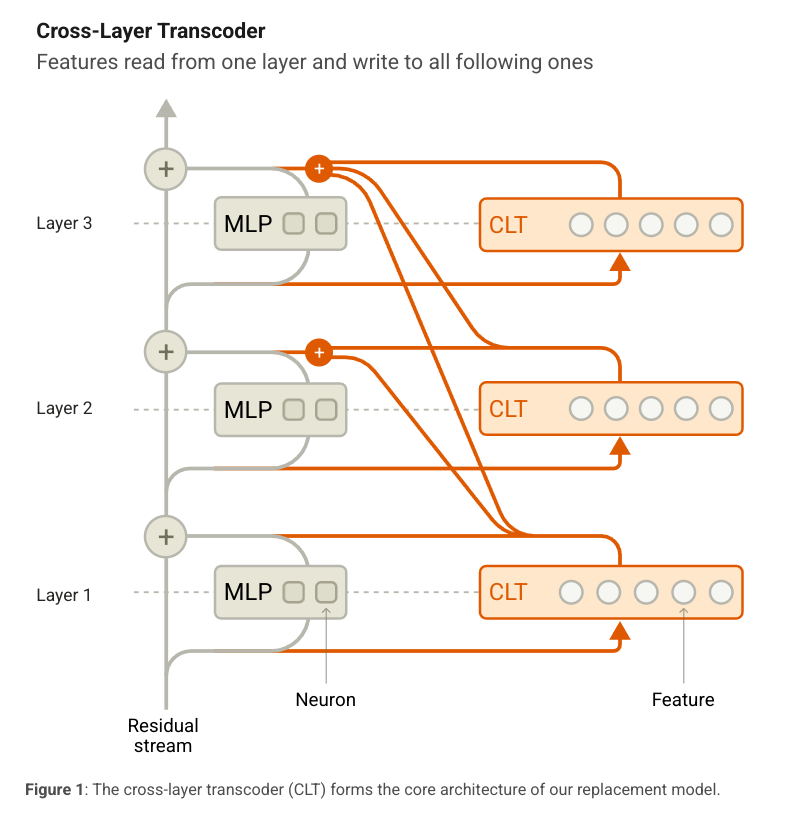
cross-layer transcoder
class RectangleFunction(autograd.Function):
@staticmethod
def forward(ctx, x):
ctx.save_for_backward(x)
return ((x > -0.5) & (x < 0.5)).float()
@staticmethod
def backward(ctx, grad_output):
(x,) = ctx.saved_tensors
grad_input = grad_output.clone()
grad_input[(x <= -0.5) | (x >= 0.5)] = 0
return grad_input
class JumpReLUFunction(autograd.Function):
@staticmethod
def forward(ctx, x, log_threshold, bandwidth):
ctx.save_for_backward(x, log_threshold, torch.tensor(bandwidth))
threshold = torch.exp(log_threshold)
return x * (x > threshold).float()
@staticmethod
def backward(ctx, grad_output):
x, log_threshold, bandwidth_tensor = ctx.saved_tensors
bandwidth = bandwidth_tensor.item()
threshold = torch.exp(log_threshold)
x_grad = (x > threshold).float() * grad_output
threshold_grad = (
-(threshold / bandwidth)
* RectangleFunction.apply((x - threshold) / bandwidth)
* grad_output
)
return x_grad, threshold_grad, None # None for bandwidth
class JumpReLU(nn.Module):
def __init__(self, feature_size, bandwidth, device='cpu'):
super(JumpReLU, self).__init__()
self.log_threshold = nn.Parameter(torch.zeros(feature_size, device=device))
self.bandwidth = bandwidth
def forward(self, x):
return JumpReLUFunction.apply(x, self.log_threshold, self.bandwidth)
class StepFunction(autograd.Function):
@staticmethod
def forward(ctx, x, log_threshold, bandwidth):
ctx.save_for_backward(x, log_threshold, torch.tensor(bandwidth))
threshold = torch.exp(log_threshold)
return (x > threshold).float()
@staticmethod
def backward(ctx, grad_output):
x, log_threshold, bandwidth_tensor = ctx.saved_tensors
bandwidth = bandwidth_tensor.item()
threshold = torch.exp(log_threshold)
x_grad = torch.zeros_like(x)
threshold_grad = (
-(1.0 / bandwidth)
* RectangleFunction.apply((x - threshold) / bandwidth)
* grad_output
)
return x_grad, threshold_grad, None # None for bandwidth
def uniform_init(tensor, limit):
with torch.no_grad():
tensor.uniform_(-limit, limit)
class CrossLayerTranscoder(nn.Module):
def __init__(self,
layer_feature_list: List[int],
d_model: int,
bandwidth: float = 1.0,
device="cpu"):
"""
layer_feature_list: e.g. [128, 256, 128, ...], length = n_layers
specifying how many features for each layer
d_model: hidden dimension of the underlying Transformer
bandwidth: controls JumpReLU partial derivatives
device: 'cpu' or 'cuda'
"""
super().__init__()
self.n_layers = len(layer_feature_list)
self.layer_feature_list = layer_feature_list
self.d_model = d_model
self.bandwidth = bandwidth
self.device = device
# (1) ENCODERS:
# We'll store a separate W_enc[i] for each layer i, shape: [layer_feature_list[i], d_model].
# We'll place them in a nn.ParameterList so we can do "W_enc[i]" in forward code.
self.W_enc = nn.ParameterList()
for i, feat_count in enumerate(layer_feature_list):
limit = 1.0 / math.sqrt(feat_count)
param = nn.Parameter(torch.empty(feat_count, d_model, device=device))
uniform_init(param, limit)
self.W_enc.append(param)
# (2) DECODERS:
# For each (src -> tgt) with src<=tgt,
# we define a matrix [layer_feature_list[src], d_model].
# We'll store them in a single nn.ParameterList, but keep track in index_map.
self.W_dec = nn.ParameterList()
self.index_map = []
dec_limit = 1.0 / math.sqrt(self.n_layers * d_model)
idx_counter = 0
for src_layer in range(self.n_layers):
row = []
src_feat = layer_feature_list[src_layer]
for tgt_layer in range(self.n_layers):
if tgt_layer >= src_layer:
dec_param = nn.Parameter(torch.empty(src_feat, d_model, device=device))
uniform_init(dec_param, dec_limit)
self.W_dec.append(dec_param)
row.append(idx_counter)
idx_counter += 1
else:
row.append(None)
self.index_map.append(row)
# (3) JumpReLUs:
# If each layer has a different #features, we can either store
# one JumpReLU for each layer, or do something simpler.
# For demonstration, we'll store one per layer:
from torch import autograd
self.jumps = nn.ModuleList()
for i, feat_count in enumerate(layer_feature_list):
self.jumps.append(JumpReLU(feat_count, bandwidth, device=device))
def forward(self, resid_streams: List[torch.Tensor]) -> List[torch.Tensor]:
"""
resid_streams[i]: shape [batch, seq, d_model], for layer i
Returns: list of length n_layers, each [batch, seq, d_model],
the reconstruction for each layer's MLP out
"""
batch_size, seq_len, _ = resid_streams[0].shape
all_activations = [] # a^ℓ for each layer
# 1) ENCODING
for i in range(self.n_layers):
x = resid_streams[i] # [batch, seq, d_model]
W_enc_mat = self.W_enc[i] # [layer_feature_list[i], d_model]
# => a_pre shape [batch, seq, layer_feature_list[i]]
a_pre = torch.einsum("bsd,nd->bsn", x, W_enc_mat)
# jump layer i
a_post = self.jumps[i](a_pre)
all_activations.append(a_post)
# 2) DECODING
# y^ℓ_hat = sum_{ℓ'<=ℓ} W_dec^(ℓ'->ℓ) * a^(ℓ')
mlp_recon = []
for tgt_layer in range(self.n_layers):
recon = torch.zeros(batch_size, seq_len, self.d_model, device=self.device)
for src_layer in range(tgt_layer+1):
dec_idx = self.index_map[src_layer][tgt_layer]
W_dec_mat = self.W_dec[dec_idx] # shape [layer_feature_list[src_layer], d_model]
a_src = all_activations[src_layer] # [batch, seq, layer_feature_list[src_layer]]
# => [batch, seq, d_model]
recon_part = torch.einsum("bsn,nd->bsd", a_src, W_dec_mat)
recon += recon_part
mlp_recon.append(recon)
return mlp_recon
def forward_with_preacts(self, resid_streams: List[torch.Tensor]
) -> Tuple[List[torch.Tensor], List[torch.Tensor]]:
"""
Same as forward(), but also returning the raw pre-activations a_pre for each layer.
"""
batch_size, seq_len, _ = resid_streams[0].shape
all_a_pre = []
all_a_post = []
for i in range(self.n_layers):
x = resid_streams[i]
W_enc_mat = self.W_enc[i]
a_pre = torch.einsum("bsd,nd->bsn", x, W_enc_mat)
a_post = self.jumps[i](a_pre)
all_a_pre.append(a_pre)
all_a_post.append(a_post)
mlp_recon = []
for tgt_layer in range(self.n_layers):
recon = torch.zeros(batch_size, seq_len, self.d_model, device=self.device)
for src_layer in range(tgt_layer+1):
dec_idx = self.index_map[src_layer][tgt_layer]
W_dec_mat = self.W_dec[dec_idx]
a_src = all_a_post[src_layer]
recon_part = torch.einsum("bsn,nd->bsd", a_src, W_dec_mat)
recon += recon_part
mlp_recon.append(recon)
return mlp_recon, all_a_pre
@classmethod
def from_hookedtransformer(cls,
hmodel,
layer_feature_list: List[int],
bandwidth=1.0,
device="cpu"):
"""
hmodel: a HookedTransformer (from transformer_lens)
layer_feature_list: e.g. [128, 256, ...], length = hmodel.cfg.n_layers
"""
L = hmodel.cfg.n_layers
d_model = hmodel.cfg.d_model
if len(layer_feature_list) != L:
raise ValueError(f"layer_feature_list must have length {L}, got {len(layer_feature_list)}")
return cls(
layer_feature_list=layer_feature_list,
d_model=d_model,
bandwidth=bandwidth,
device=device
)
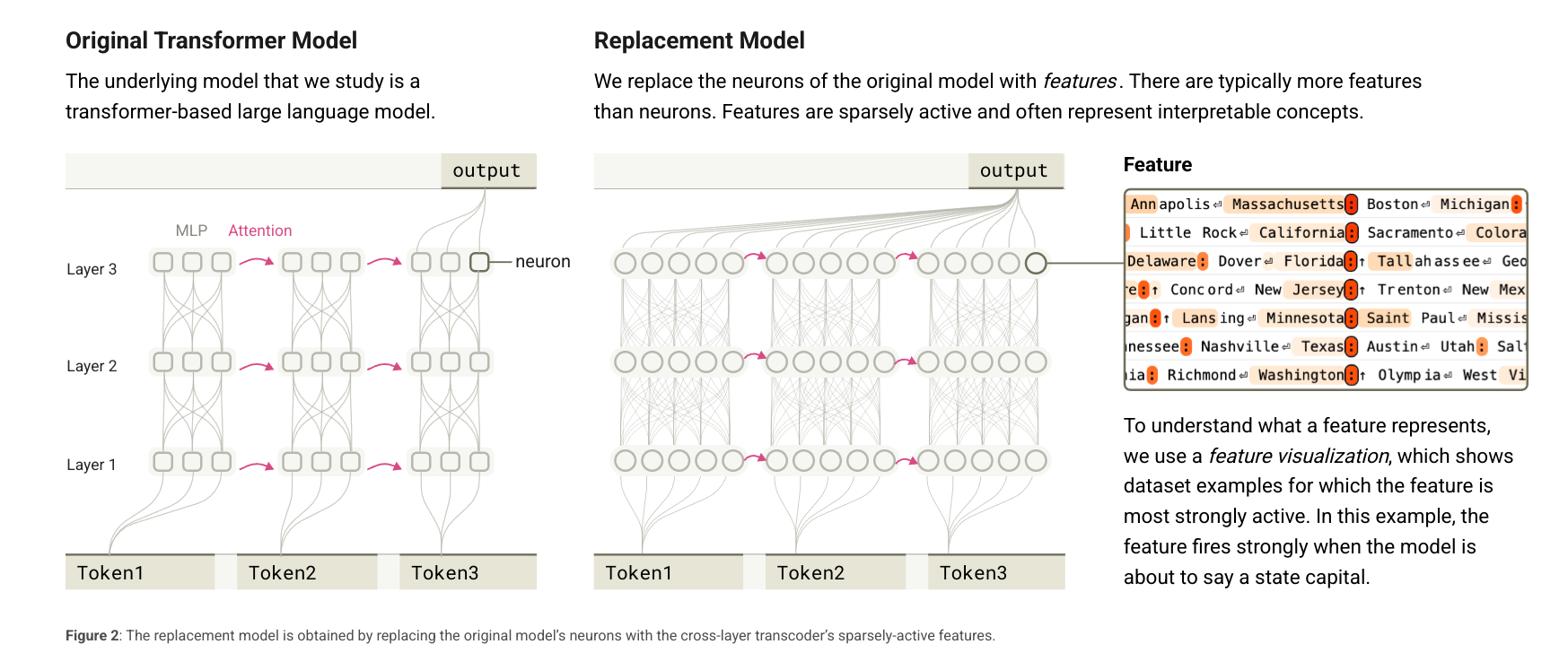
so a cross-layer transcoder is a module that takes in a list of residual streams and returns a list of reconstructed residual streams. as the paper says, it’s goal is to reconstruct the activations of the MLPs in the model.
to re-explain an already well explained concept (from the blog):
- each feature reads from the residual stream of the layer it’s in using a linear encoder
- a given layer’s features helps reconstruct the residual stream of all layers below it using a linear decoder
- features are trained jointly thus the output of an MLP is reconstructed from the features of all the layers below it
training the cross-layer transcoder
to train the CLT, there are two loss functions:
- a reconstruction loss summed over all layers
- a sparsity penalty summed over all layers
the reconstruction loss is obvious, we want to reconstruct the MLP outputs from the features, and we use MSE to do this.
the sparsity penalty is a bit more complex, but the idea is we want to encourage the model to use as few features as possible to reconstruct the MLP outputs
- this promotes interpretability: without these constraints, the model can many features to reconstruct the MLP outputs, which makes it harder attribute specific behavior to a single feature.
- networks exibit polysemanticity, where a single feature can have multiple meanings. this sparsity constraint allows us to monosemanticity, where a single feature has a single meaning.
- reducing noise in the eventual attribution graphs: having too many active features can lead to incredibly dense attribution graphs, which are harder to analyze.
def advanced_sparsity_loss(
clt: CrossLayerTranscoder,
preacts_list: List[torch.Tensor],
c: float = 1.0,
lambda_spars: float = 1e-3
):
"""
Example: sum_{src_layer, i} tanh( c * ||W_dec_{i}|| * mean(a_pre) )
We'll do a simplified version, ignoring multi-step or partial layering.
"""
device = preacts_list[0].device
L = clt.n_layers
penalty = torch.zeros((), device=device)
# for each layer i
for src_layer in range(L):
a_pre = preacts_list[src_layer] # shape [batch, seq, n_feat_this_layer]
# average activation across batch, seq => [n_feat_this_layer]
a_mean = a_pre.mean(dim=(0,1))
# for each tgt_layer >= src_layer
for tgt_layer in range(src_layer, L):
dec_idx = clt.index_map[src_layer][tgt_layer]
W_dec_mat = clt.W_dec[dec_idx] # shape [layer_feature_list[src_layer], d_model]
dec_norm = W_dec_mat.norm(dim=1) # shape [layer_feature_list[src_layer]]
raw_vals = c * dec_norm * a_mean
penalty_layer = torch.tanh(raw_vals).sum()
penalty += penalty_layer
return lambda_spars * penalty
def get_sparsity_scale(current_step, total_steps, lambda_final):
# linear ramp from 0 to lambda_final across total_steps
scale = min(1.0, current_step / (total_steps - 1))
return scale * lambda_final
def sum_of_relu_neg(preacts_list):
"""
preacts_list: a list of Tensors, each shape [batch, seq, n_features],
containing the pre-activation values for each layer.
Returns a scalar that is the sum over all layers, batches, and feature dims of
ReLU(-preactivation).
"""
total_loss = torch.tensor(0.0, device=preacts_list[0].device)
for a_pre in preacts_list:
# shape: [batch, seq, n_features]
negvals = torch.relu(-a_pre) # ReLU(-x) = max(0, -x)
total_loss += negvals.sum()
return total_loss
def train_clt(
clt,
dataloader,
num_epochs,
lambda_spars_final=1e-3,
preact_loss_coef=3e-6,
total_steps=None
):
"""
clt: CrossLayerTranscoder module with JumpReLU threshold=0.03, uniform init, etc.
dataloader: yields (resid_list, mlp_list) already normalized
"""
optimizer = torch.optim.Adam(clt.parameters(), lr=1e-4)
if total_steps is None:
total_steps = num_epochs * len(dataloader)
global_step = 0
for epoch in range(num_epochs):
print(f"Epoch {epoch} of {num_epochs}")
for batch_idx, (resid_batch_list, mlp_batch_list) in enumerate(dataloader):
print(f"Batch {batch_idx} of {len(dataloader)}")
batch_size = resid_batch_list[0].shape[0]
# to GPU
# remember to ensure these are normalized per token
for l in range(len(resid_batch_list)):
# need this for weird batching error
resid_batch_list[l] = resid_batch_list[l].to(clt.device)[0]
mlp_batch_list[l] = mlp_batch_list[l].to(clt.device)[0]
# forward pass => recon_list
recon_list, preactivations = clt.forward_with_preacts(resid_batch_list)
# ^ maybe you define forward_with_preacts() to return
# the list of preactivation a^ell and the final recon.
L_preact = 3e-6 * sum_of_relu_neg(preactivations)
# MSE
mse = torch.tensor(0.0, device=clt.device)
for i in range(len(recon_list)):
diff = recon_list[i] - mlp_batch_list[i]
mse += diff.pow(2).mean()
# advanced sparsity
current_lambda_spars = get_sparsity_scale(
global_step, total_steps, lambda_spars_final
)
L_spars = advanced_sparsity_loss(
clt,
preactivations, # the list of shape [batch, seq, n_features] for each layer
c=1.0,
lambda_spars=current_lambda_spars
)
# preact loss
# preactivations is a list of shape [batch, n_features] for each layer
total_loss = mse + L_spars + L_preact
optimizer.zero_grad()
total_loss.backward()
optimizer.step()
global_step += 1
print(f"Epoch {epoch} done. last total_loss={total_loss.item():.4f} MSE={mse.item():.4f}")
dataset = CLTHiddenStateDataset("clt_data")
dataloader = DataLoader(dataset, batch_size=1, shuffle=True, num_workers=4)
model_name = "gpt2-small"
hmodel = HookedTransformer.from_pretrained(model_name)
layer_feature_list = [128]*hmodel.cfg.n_layers # or something custom
clt = CrossLayerTranscoder.from_hookedtransformer(
hmodel, layer_feature_list, bandwidth=1.0, device="cuda"
)
train_clt(clt, dataloader, 2)
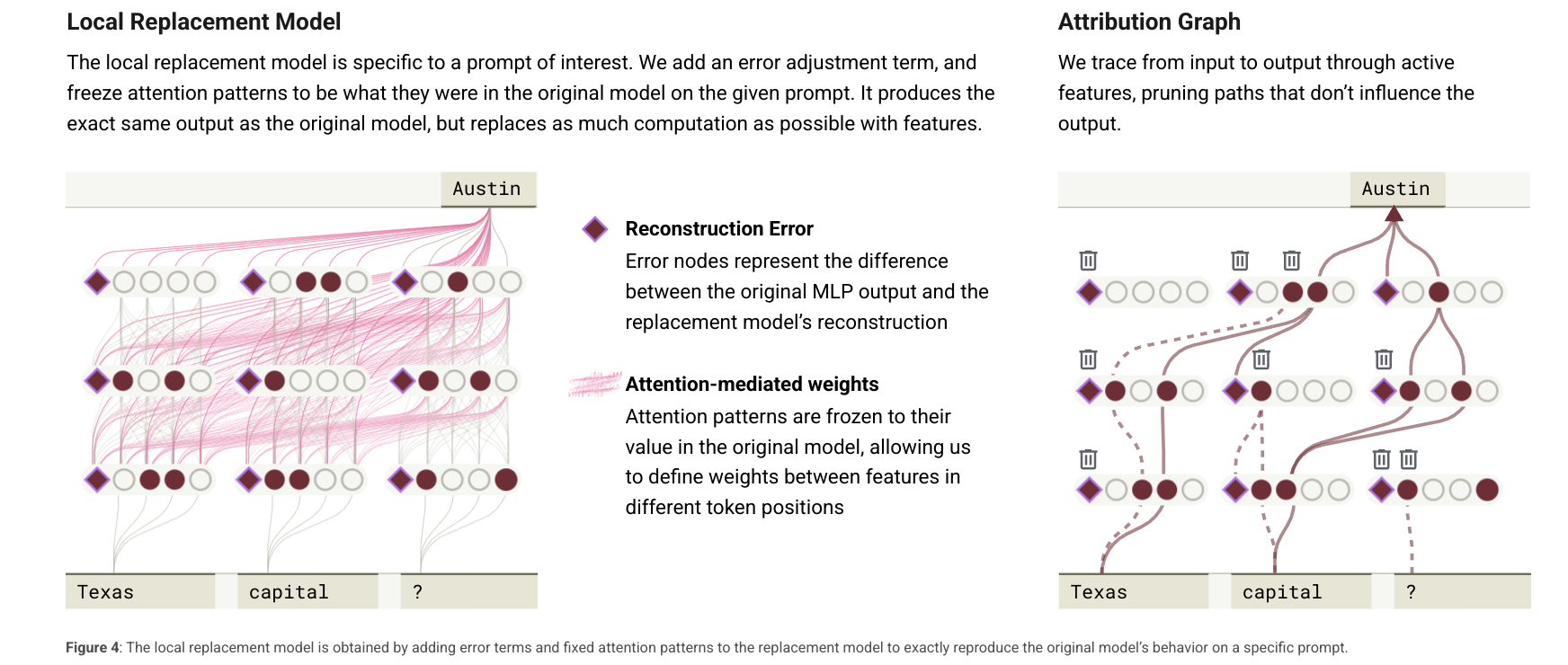
replacement modeling
basically, a replacement model is the swapping out of the MLPs in the model with the CLT while also providing an error correction term to account for the error in the reconstruction.
the way anthropic frames this is that this basically re-writes the underlying model with sparser, more interpretable units.
the way anthropic describes how we should view the local replacement model is such:
it’s a fully connected neural network spanning across tokens that allow us to do interpretability at research
- effectively, it’s a graph.
- it’s the union of CLT features active at every token position.
- Its weights are the summed interactions over all the linear paths from one feature to another
def get_layer_index_from_hook(hook_name: str):
"""
A helper that extracts the integer layer index from a name like
'blocks.3.hook_mlp_out' or 'blocks.3.hook_resid_mid', etc.
Returns None if not recognized.
"""
# e.g. "blocks.3.hook_mlp_out" -> 3
# naive approach:
if "blocks." not in hook_name:
return None
try:
after_blocks = hook_name.split("blocks.")[1]
i_str = after_blocks.split(".")[0] # e.g. "3"
return int(i_str)
except:
return None
class LocalReplacementModel(nn.Module):
"""
A local replacement model that:
1) Uses `base_transformer` for the general structure,
2) Freezes LN outputs & attention patterns with pre-recorded data,
3) Replaces each MLP with CLT + error corrections,
4) Records the cross-layer-transcoder's feature activations in self.clt_activations.
"""
def __init__(
self,
base_transformer: HookedTransformer,
clt: "CrossLayerTranscoder",
error_corrections: Dict[str, torch.Tensor],
ln_scales: Dict[str, torch.Tensor],
attn_patterns: Dict[str, torch.Tensor],
device="cuda"
):
super().__init__()
self.base = base_transformer
self.clt = clt
self.error_corrections = error_corrections # e.g. { "blocks.3.hook_mlp_out": tensor(...) }
self.ln_scales = ln_scales # e.g. { "blocks.3.ln1.hook_normalized": tensor(...) }
self.attn_patterns = attn_patterns # e.g. { "blocks.3.attn.hook_pattern": tensor(...) }
self.device = device
# We'll store the MLP inputs for each layer (the "hook_mlp_in" values)
self.mlp_inputs = [None]*clt.n_layers
# We'll also store the cross-layer feature activations for each layer
# (shape [batch, seq, n_features_of_layer]) after we run the MLP-out hook
self.clt_activations = [None]*clt.n_layers
def forward(self, tokens: torch.Tensor) -> torch.Tensor:
"""
1. Freeze LN by injecting recorded LN outputs.
2. Freeze QK by injecting stored attention patterns.
3. Replace MLP with CLT + error corrections.
4. Store CLT feature activations in self.clt_activations.
"""
# ===== Define hooking functions =====
def freeze_ln_hook(activation, hook):
"""
Hook on e.g. "blocks.0.ln1.hook_normalized":
We replace the LN output with our pre-saved LN output (from an earlier pass).
"""
name = hook.name
if name in self.ln_scales:
return self.ln_scales[name].to(self.device)
return activation
def freeze_attn_pattern_hook(activation, hook):
"""
Hook on e.g. "blocks.0.attn.hook_pattern":
We replace the QK-softmax pattern with pre-saved patterns.
"""
name = hook.name
if name in self.attn_patterns:
return self.attn_patterns[name].to(self.device)
return activation
def mlp_in_hook(activation, hook):
"""
E.g. "blocks.3.hook_mlp_in" => store for layer 3
"""
layer_idx = get_layer_index_from_hook(hook.name)
if layer_idx is not None:
self.mlp_inputs[layer_idx] = activation.detach().clone()
return activation
def mlp_out_hook(activation, hook):
"""
E.g. "blocks.3.hook_mlp_out" => we do:
1) feed self.mlp_inputs[L] into the CLT for layer L,
2) add error correction,
3) also fill self.clt_activations.
"""
layer_idx = get_layer_index_from_hook(hook.name)
if layer_idx is None:
return activation
x = self.mlp_inputs[layer_idx]
if x is None:
return activation # fallback
# We'll feed "dummy_resids" so that only layer_idx's input is nonzero
dummy_resids = []
for i in range(self.clt.n_layers):
if i == layer_idx:
dummy_resids.append(x)
else:
dummy_resids.append(torch.zeros_like(x))
# We want the "pre-activations" a_pre from each layer,
# so we use forward_with_preacts:
mlp_recon_list, all_a_post = self.clt.forward_with_preacts(dummy_resids)
# mlp_recon_list => list of length n_layers, each shape [batch, seq, d_model]
# all_a_post => list of length n_layers, each shape [batch, seq, n_feat(i)]
# The reconstructed MLP-out for this layer
recon_layer = mlp_recon_list[layer_idx]
# Let's store the cross-layer feature activations for *all* layers
# (each a_post is shape [batch, seq, #features for that layer])
for i, feat_acts in enumerate(all_a_post):
self.clt_activations[i] = feat_acts.detach().clone()
# Then add the "error correction," if any
err_key = hook.name # e.g. "blocks.3.hook_mlp_out"
if err_key in self.error_corrections:
e = self.error_corrections[err_key].to(self.device)
recon_layer = recon_layer + e
return recon_layer
# ===== Attach the hooks =====
hooks = []
# Freeze LN
for i in range(self.base.cfg.n_layers):
ln_name = f"blocks.{i}.ln1.hook_normalized"
if ln_name in self.base.hook_dict:
h_ln = self.base.add_hook(ln_name, freeze_ln_hook, "fwd")
hooks.append(h_ln)
# Freeze attention pattern
for i in range(self.base.cfg.n_layers):
attn_name = f"blocks.{i}.attn.hook_pattern"
if attn_name in self.base.hook_dict:
h_attn = self.base.add_hook(attn_name, freeze_attn_pattern_hook, "fwd")
hooks.append(h_attn)
# Intercept MLP in/out
for i in range(self.clt.n_layers):
in_name = f"blocks.{i}.hook_mlp_in"
out_name = f"blocks.{i}.hook_mlp_out"
if in_name in self.base.hook_dict:
hi = self.base.add_hook(in_name, mlp_in_hook, "fwd")
hooks.append(hi)
if out_name in self.base.hook_dict:
ho = self.base.add_hook(out_name, mlp_out_hook, "fwd")
hooks.append(ho)
# ===== Run forward pass =====
logits = self.base(tokens)
# remove hooks after finishing
for h in hooks:
if h is not None:
h.remove()
return logits
def build_local_replacement_model_with_cache(
base_model: HookedTransformer,
clt: CrossLayerTranscoder,
prompt: str,
device="cuda"
):
"""
1) forward pass on base_model, store LN outputs, attention patterns, MLP in/out
2) compute error corrections
3) return a LocalReplacementModel that re-uses LN & attn
and MLP is replaced with CLT+error
"""
layer_count = base_model.cfg.n_layers
tokens = base_model.to_tokens(prompt, prepend_bos=True).to(device)
# -- Use a filter that picks up the hook names we want. --
# We want LN outputs, attn pattern, mlp_in, mlp_out
# For LN: "blocks.{i}.ln1.hook_normalized"
# For attn: "blocks.{i}.attn.hook_pattern"
# For MLP in/out: "blocks.{i}.hook_mlp_in" / "blocks.{i}.hook_mlp_out"
def activation_filter(name: str):
# Return True if we want to store this hook in the cache
# Return False otherwise
if ".ln1.hook_normalized" in name:
return True
if ".attn.hook_pattern" in name:
return True
if ".hook_mlp_in" in name:
return True
if ".hook_mlp_out" in name:
return True
return False
# run_with_cache returns (logits, cache).
# cache is a HookedTransformerCache containing the stored activations
logits, cache = base_model.run_with_cache(
tokens,
return_type="logits", # or "none" if you don't need final logits
names_filter=activation_filter
)
ln_scales = {}
attn_patterns = {}
mlp_in_cache = {}
mlp_out_cache = {}
# read from cache for each layer's LN out, attn pattern, MLP in/out
for i in range(layer_count):
ln_key = f"blocks.{i}.ln1.hook_normalized"
attn_key = f"blocks.{i}.attn.hook_pattern"
in_key = f"blocks.{i}.hook_mlp_in"
out_key = f"blocks.{i}.hook_mlp_out"
if ln_key in cache:
ln_scales[ln_key] = cache[ln_key].detach().clone()
if attn_key in cache:
attn_patterns[attn_key] = cache[attn_key].detach().clone()
if in_key in cache:
mlp_in_cache[in_key] = cache[in_key].detach().clone()
if out_key in cache:
mlp_out_cache[out_key] = cache[out_key].detach().clone()
# 2) Build error corrections by comparing the model's MLP out to the CLT recon
error_corrections = {}
for i in range(clt.n_layers):
layer_in_name = f"blocks.{i}.hook_mlp_in"
layer_out_name = f"blocks.{i}.hook_mlp_out"
# Only do this if we actually have MLP in/out for that layer
if layer_in_name in mlp_in_cache and layer_out_name in mlp_out_cache:
mlp_in = mlp_in_cache[layer_in_name].to(device)
mlp_out_true = mlp_out_cache[layer_out_name].to(device)
# minimal forward in CLT for just this layer
dummy_inputs = [torch.zeros_like(mlp_in) for _ in range(clt.n_layers)]
dummy_inputs[i] = mlp_in
clt_outputs = clt(dummy_inputs) # list of shape [n_layers]
clt_layer_out = clt_outputs[i] # [batch, seq, d_model]
diff = mlp_out_true - clt_layer_out
error_corrections[layer_out_name] = diff.detach().clone()
# 3) Build the final "frozen" local model
local_model = LocalReplacementModel(
base_transformer=base_model,
clt=clt,
error_corrections=error_corrections,
ln_scales=ln_scales,
attn_patterns=attn_patterns,
device=device
)
return local_model
Attribution Graphs
these are representations of how different computational components (specifically the interpretable features) contribute to the final output of the model.
graphs contain 4 nodes:
- output: the final output tokens of the model
- input: the input embedding tokens to the model
- intermediate: clt features at each prompt token
- error: the remaining output unexplained by the CLT
edges represent linear attributions; they originate from the input node and end at the output node and indicate a direct, linear influence from one node to another. the idea is that the activation of any feature (or token) is decomposed into the sum of its incoming contributions from these edges, allowing a clear, linear causal interpretation.
the graph is constructed by backward attribution via jacobians.
so we want to decompose the output into the sum of contributions from earlier components. because the influence is spread over many different paths, using backward jacobians allows one to quantify the sensitivity of the target’s pre-activation to changes in the source’s activation.
< explain math/gradients >
for every target t in our graph, we inject its corresponding input vector into the residual stream at the appropriate layer/token position. we then perform a backward pass on the underlying model with the following modifications: 1) stop-gradients are inserted into the non-linear parts of the model and 2) frozen attention patterns are used so that the only way the input can affect the output is through the linear paths in the graph. for any source node, its contribution is computed as a dot product between its decoder vector and the gradient signal that flows from the target to the source. lastly, the result is scaled by the activation of the source node.
by computing these backward jacobians, we effectively obtain a set of linear influence weights that hell us how much each feature contributes to the target. by doing this, you can effectively map out a circuit (in the mech interp sense).
anthropic also describes a graph pruning algorithm that they describe with the following pseudocode:
function compute_normalized_adjacency_matrix(graph):
# Convert graph to adjacency matrix A
# A[j, i] = weight from i to j (note the transposition)
A = convert_graph_to_adjacency_matrix(graph)
A = absolute_value(A)
# Normalize each row to sum to 1
row_sums = sum(A, axis=1)
row_sums = maximum(row_sums, 1e-8) # Avoid division by zero
A = diagonal_matrix(1/row_sums) @ A
return A
function prune_nodes_by_indirect_influence(graph, threshold):
A = compute_normalized_adjacency_matrix(graph)
# Calculate the indirect influence matrix: B = (I - A)^-1 - I
# This is a more efficient way to compute A + A^2 + A^3 …
B = inverse(identity_matrix(size=A.shape[0]) - A) - identity_matrix(size=A.shape[0])
# Get weights for logit nodes.
# This is 0 if a node is a non-logit node and equal to the probability for logit nodes
logit_weights = get_logit_weights(graph)
# Calculate influence on logit nodes for each node
influence_on_logits = matrix_multiply(B, logit_weights)
# Sort nodes by influence
sorted_node_indices = argsort(influence_on_logits, descending=True)
# Calculate cumulative influence
cumulative_influence = cumulative_sum(
influence_on_logits[sorted_node_indices]) / sum(influence_on_logits)
# Keep nodes with cumulative influence up to threshold
nodes_to_keep = cumulative_influence <= threshold
# Create new graph with only kept nodes and their edges
return create_subgraph(graph, nodes_to_keep)
# Edge pruning by thresholded influence
function prune_edges_by_thresholded_influence(graph, threshold):
# Get normalized adjacency matrix
A = compute_normalized_adjacency_matrix(graph)
# Calculate influence matrix (as before)
B = estimate_indirect_influence(A)
# Get logit node weights (as before)
logit_weights = get_logit_weights(graph)
# Calculate node scores (influence on logits)
node_score = matrix_multiply(B, logit_weights)
# Edge score is weighted by the logit influence of the target node
edge_score = A * node_score[:, None]
# Calculate edges to keep based on thresholded cumulative score
sorted_edges = sort(edge_score.flatten(), descending=True)
cumulative_score = cumulative_sum(sorted_edges) / sum(sorted_edges)
threshold_index = index_where(cumulative_score >= threshold)
edge_mask = edge_score >= sorted_edges[threshold_index]
# Create new graph with pruned adjacency matrix
pruned_adjacency = A * edge_mask
return create_subgraph_from_adjacency(graph, pruned_adjacency)
my implementation of the attribution graph code is as follows:
class NodeType:
EMBEDDING = "embedding"
FEATURE = "feature"
ERROR = "error"
LOGIT = "logit"
class AttributionNode:
"""
A node in the attribution graph, e.g. a single feature in some layer,
or an embedding node, or a logit node, etc.
"""
def __init__(
self,
node_type: str,
name: str,
layer_idx: int = None,
context_pos: int = None,
logit_index: int = None
):
self.node_type = node_type
self.name = name
# Set layer_idx to a sentinel if not provided
if layer_idx is None:
if node_type == NodeType.EMBEDDING:
self.layer_idx = -1
elif node_type == NodeType.LOGIT:
self.layer_idx = 9999
else:
self.layer_idx = 0
else:
self.layer_idx = layer_idx
self.context_pos = context_pos
self.logit_index = logit_index
# Storing these as placeholders
self.activation = None
self.output_vector = None
self.input_vector = None
# Must be an integer for feature nodes:
self.feature_index = None # we must fill this in ourselves
# We'll store an integer ID in the graph
self.id = None
def __repr__(self):
return f"Node<{self.node_type}:{self.name}:{self.id}>"
class AttributionGraph:
"""
A container for all nodes and edges in the attribution graph.
"""
def __init__(self):
self.nodes = []
self.edges = {} # adjacency: edges[u] = list of (v, weight)
def add_node(self, node: AttributionNode):
node.id = len(self.nodes)
self.nodes.append(node)
self.edges[node.id] = []
return node.id
def add_edge(self, src_id: int, tgt_id: int, weight: float):
self.edges[src_id].append((tgt_id, weight))
def get_num_nodes(self):
return len(self.nodes)
def build_graph_nodes(
local_model, # e.g. a LocalReplacementModel
prompt: str,
top_k=3,
feature_threshold=0.01
):
"""
1) Forward pass => local_model.clt_activations is populated
2) Build an AttributionGraph
3) Add logit nodes for top_k final predictions
4) Add feature nodes for each cross-layer feature whose activation > threshold
"""
G = AttributionGraph()
tokens = local_model.base.to_tokens(prompt, prepend_bos=True).to(local_model.device)
with torch.no_grad():
logits = local_model(tokens) # fills local_model.clt_activations
# Add top-k logit nodes
final_logits = logits[0, -1, :]
probs = F.softmax(final_logits, dim=-1)
top_vals, top_inds = torch.topk(final_logits, top_k)
logit_node_ids = []
for rank in range(top_k):
tok_id = top_inds[rank].item()
p = probs[tok_id].item()
node = AttributionNode(
node_type=NodeType.LOGIT,
name=f"logit_{tok_id} (p={p:.3f})",
layer_idx=None, # sets layer_idx=9999
context_pos=tokens.shape[1]-1, # last position
logit_index=tok_id
)
nid = G.add_node(node)
logit_node_ids.append(nid)
# Create feature nodes from local_model.clt_activations
feature_nodes = {}
for i in range(local_model.clt.n_layers):
acts_i = local_model.clt_activations[i] # shape [1, seq, n_feat_i]
if acts_i is None:
continue
acts_i = acts_i[0] # shape [seq, n_feat_i] if batch=1
seq_len, n_feat_i = acts_i.shape
for pos in range(seq_len):
for feat_j in range(n_feat_i):
val = acts_i[pos, feat_j].item()
if val > feature_threshold:
# Build a feature node => layer i, position pos, feature index feat_j
node = AttributionNode(
node_type=NodeType.FEATURE,
name=f"feat_L{i}_f{feat_j}_pos{pos}",
layer_idx=i,
context_pos=pos,
logit_index=None
)
node.activation = val
# Key fix: assign the integer feature index
node.feature_index = feat_j # <--- CRUCIAL
node_id = G.add_node(node)
feature_nodes[(i, pos, feat_j)] = node_id
cache_dict = {
"logit_node_ids": logit_node_ids,
"feature_nodes": feature_nodes
}
return G, cache_dict
def compute_logit_injection_vector(local_model, vocab_idx):
"""
Suppose you do 'logits = W_U * final_resid'. Then the gradient for
(logit[vocab_idx] - mean(logit)) w.r.t. final_resid is:
injection_vec = W_U[vocab_idx] - average(W_U)
or something along these lines.
"""
W_U = local_model.base.W_U # e.g. unembedding weights shape [d_model, vocab_size]
d_model, vocab_size = W_U.shape
# minimal example:
w_target = W_U[:, vocab_idx] # shape [d_model]
w_mean = W_U.mean(dim=1) # shape [d_model]
injection_vec = (w_target - w_mean)
return injection_vec
def run_backward_with_injection(
local_model: LocalReplacementModel,
injection_vec: torch.Tensor,
layer_idx: int,
real_tokens: torch.Tensor,
token_pos: int = None,
freeze_ln=True,
freeze_attn=True,
):
"""
Runs a backward pass in 'local_model' (a LocalReplacementModel) with an
'injection_vec' placed into the chosen layer (layer_idx) residual stream.
We now pass 'real_tokens' so that the dimension of the residual
matches the dimension we used in the normal forward pass.
Args:
local_model: The local replacement model (with LN & attn patterns frozen).
injection_vec: a Tensor of shape [d_model] or [batch, seq, d_model],
specifying the "gradient signal" we want to inject.
layer_idx: which layer's residual we inject into.
real_tokens: the actual token IDs used in the normal forward pass
(must have the same shape as originally used by local_model).
token_pos: which token position to inject into. If None, we broadcast across all positions.
freeze_ln: whether to freeze LN denominators in the backward pass.
freeze_attn: whether to freeze QK patterns in the backward pass.
Returns:
grad_dict: a dictionary of shape {(layer_idx, pos): residual_grad_vector}
containing the gradient w.r.t. each layer's residual stream after injection.
"""
# (A) clamp layer_idx if needed
nL = local_model.base.cfg.n_layers
if layer_idx >= nL:
# Perhaps for logit nodes, set it to nL - 1
layer_idx = nL - 1
# 1) Zero out old gradients
local_model.zero_grad(set_to_none=True)
# 2) Optionally freeze LN or QK params so they do not accumulate gradient
# This ensures that no gradient accumulates in LN or QK parameters.
# Just be mindful that some models might have LN named differently (ln_f or ln_final).
if freeze_ln or freeze_attn:
for name, param in local_model.base.named_parameters():
if freeze_ln and ("ln" in name):
param.requires_grad_(False)
if freeze_attn and (".W_Q" in name or ".W_K" in name):
param.requires_grad_(False)
# 3) Hook the chosen layer's residual via a forward pass with the *real tokens*
storage = {}
def store_resid_hook(resid, hook):
# resid shape is [batch, seq, d_model]
storage["resid"] = resid
return resid
# By default for GPT-2 style, the "pre-MLP residual" is often "blocks.{layer_idx}.hook_resid_mid"
# or "blocks.{layer_idx}.hook_resid_pre". For GPT-2 from TransformerLens, it's typically:
# "blocks.{layer_idx}.hook_resid_mid"
# but the user's code might vary. We'll try "hook_resid_mid" here:
resid_name = f"blocks.{layer_idx}.hook_resid_mid"
if resid_name not in local_model.base.hook_dict:
# fallback or raise error
# e.g. check if "hook_resid_post" is present
alt_name = f"blocks.{layer_idx}.hook_resid_post"
if alt_name in local_model.base.hook_dict:
resid_name = alt_name
else:
raise KeyError(f"No valid mid/post resid hook for layer {layer_idx}")
# register the forward hook
handle = local_model.base.add_hook(resid_name, store_resid_hook, "fwd")
with torch.enable_grad():
# Perform the forward pass with the REAL tokens so shapes match
_ = local_model(real_tokens)
if handle is not None:
handle.remove()
if "resid" not in storage:
raise ValueError(
f"Could not find residual for layer {layer_idx} - check your "
f"hook name (is it 'hook_resid_mid' or 'hook_resid_pre'?)"
)
resid_L = storage["resid"] # shape [batch, seq, d_model]
# We want gradient to flow from the dummy loss -> resid_L
# 4) Construct the dummy loss
# If injection_vec is [d_model], we broadcast across [batch, seq, d_model].
shape = resid_L.shape
if injection_vec.dim() == 1 and injection_vec.shape[0] == shape[-1]:
expanded_injection = injection_vec.view(1, 1, -1).expand_as(resid_L)
elif injection_vec.shape == shape:
expanded_injection = injection_vec
else:
raise ValueError(f"injection_vec shape mismatch: got {injection_vec.shape}, "
f"but resid_L is {shape}.")
if token_pos is not None:
# zero out gradient for all positions except 'token_pos'
mask = torch.zeros_like(resid_L)
mask[:, token_pos, :] = 1.0
expanded_injection = expanded_injection * mask
dummy_loss = (resid_L * expanded_injection).sum()
dummy_loss.backward(retain_graph=True)
# 5) gather gradient in a dictionary
grad_dict = {}
# For demonstration, we only store the gradient for exactly (layer_idx, token_pos)
# If you want all positions, you can store them all.
# We'll do a single key => shape [batch, seq, d_model]
if resid_L.grad is not None:
grad_dict[(layer_idx, "all")] = resid_L.grad.detach().cpu()
else:
grad_dict[(layer_idx, "all")] = None
return grad_dict
def is_upstream_layer(src_node, tgt_node):
"""
Example helper: returns True if src_node is strictly earlier in
(layer, position) than tgt_node. You can define your own logic for
"which nodes can feed into which" in your graph.
"""
# e.g. if src_node.layer_idx < tgt_node.layer_idx, or if equal layer but earlier tokenpos
# etc. We'll just do a dummy check:
return (src_node.layer_idx < tgt_node.layer_idx) or (
src_node.layer_idx == tgt_node.layer_idx
and src_node.context_pos <= tgt_node.context_pos
)
def get_token_embedding(token_id, local_model):
"""
Suppose your model has a wte (word token embed) of shape [vocab_size, d_model].
Then the embedding is wte[token_id].
"""
return local_model.base.embed[token_id] # for example
def compute_direct_edges_for_node(
G,
node_id: int,
local_model: nn.Module,
tokens: torch.Tensor,
freeze_ln=True,
freeze_attn=True,
epsilon=1e-8,
):
"""
For a given node in the attribution graph, we compute the direct edges from
*all* upstream nodes in G to `node_id`. We do a custom backward pass in the
local replacement model, with LN denominators and QK patterns frozen (stop-grad).
Pseudocode steps:
1) Build the injection vector for the `node_id`—this depends on whether it's a
Feature node, Logit node, etc.
2) Insert that injection vector into the residual stream at the correct layer
(or final-layer residual for a logit).
3) Run a custom backward pass that accumulates `.grad` in the model's residual
streams (and zero in LN denominators, etc.).
4) For each source node in G, compute the direct edge weight using either:
w = source_activation * sum_{k} [ W_dec^{(source)}^T * grad_for_layer_k * W_enc^{(target)} ]
or for embeddings / error nodes, a direct dot-product with the residual grad.
5) Add edge to G if abs(weight) > threshold.
"""
node = G.nodes[node_id]
node_type = node.node_type
# ---------------------------------------------------------------------
# 1) Build the injection vector for the "target node"
# ---------------------------------------------------------------------
if node_type == NodeType.LOGIT:
# For a logit node, commonly we do "gradient w.r.t. (logit_tok - mean_logit)".
target_logit_idx = node.logit_index # e.g. ID in vocab
# Suppose we have the final-layer residual dimension = d_model
# We make an injection vector of shape [d_model], or [1, d_model]
injection_vec = compute_logit_injection_vector(local_model, target_logit_idx)
elif node_type == NodeType.FEATURE:
# For a feature node, we typically want to inject that feature's input vector
# into the residual stream at the layer it reads from. That is:
# v_in^L = W_enc[featureID] (the encoder weights)
# For cross-layer transcoders, each feature has an encoder layer. We assume
# node stores e.g. node.layer_idx, node.feature_idx, etc.
layer_idx = node.layer_idx
feat_idx = node.feature_idx # e.g. index in that layer's set of features
injection_vec = local_model.clt.W_enc[layer_idx][feat_idx].detach().clone()
elif node_type == NodeType.ERROR:
# For error nodes, we might define injection_vec as the error node's
# "output vector" in the residual stream.
# e.g. v_out = MLP_out_true - CLT_out. We'll just pretend we have it stored:
injection_vec = node.output_vector.detach().clone()
elif node_type == NodeType.EMBEDDING:
# For an embedding node, we can do something like:
token_embed = get_token_embedding(node.token_id, local_model)
injection_vec = token_embed.detach().clone()
else:
raise ValueError(f"Unknown node_type = {node_type}")
# We'll reshape to [d_model] or [1,d_model] if needed
injection_vec = injection_vec.detach().view(-1)
injection_vec.requires_grad_(False)
# ---------------------------------------------------------------------
# 2) Insert the injection vector into the residual stream
# We'll do a custom backward pass that sets MLP out, LN denominators, QK patterns
# to no_grad or zero_grad, etc.
# ---------------------------------------------------------------------
# For example, if the node is a LOGIT node, we put injection_vec at the final-layer residual
# If it's a FEATURE in layer L, we put the injection_vec in the residual of layer L
# We'll store them in a 'tensor' that the model sees as the backward pass "incoming gradient".
# We'll define a helper function. In a real code, you'd define a specialized
# "run_backward_with_injection" that does your custom hooking logic:
grad_dict = run_backward_with_injection(
local_model=local_model,
injection_vec=injection_vec,
layer_idx=node.layer_idx if node_type == NodeType.FEATURE else local_model.base.cfg.n_layers,
# ^ if logit or error or embedding, we might set this to final layer
real_tokens=tokens,
freeze_ln=freeze_ln,
freeze_attn=freeze_attn,
)
# grad_dict: Suppose it returns a dict mapping {(layer_idx, token_pos): residual_grad_tensor}
# or something similar for each layer. You may also want the raw MLP-input grads in
# your local replacement model or the partial derivatives w.r.t. CLT decoders, etc.
# ---------------------------------------------------------------------
# 3) For each source node, compute direct edge weight
# ---------------------------------------------------------------------
# We'll show how to do it for a "feature" source node (the typical case).
# We'll also handle "embedding" and "error" node. (They differ because
# features have to multiply by their activation in this prompt.)
# We'll do a loop over all nodes in G. In practice, you might only want to
# handle nodes at earlier layers, or earlier token positions, etc.
for src_id, src_node in enumerate(G.nodes):
if src_id == node_id:
continue
if not is_upstream_layer(src_node, node): # you might have logic to skip obviously irrelevant nodes
continue
w = 0.0
if src_node.node_type == NodeType.FEATURE:
# The formula from the text is roughly:
#
# A_{s -> t} = a_s * sum_{ℓ in [src_layer..(tgt_layer-1)]} [
# W_dec(src_feat)ᵀ * grad[ℓ] * W_enc(tgt_feat)
# ]
#
# But we already have "grad[ℓ] * W_enc(tgt_feat)" from the injection pass,
# plus we can do "W_dec(src_feat)ᵀ . that" ...
# We'll do it more explicitly in code:
src_activation = src_node.activation # a_s from the prompt
s_layer = src_node.layer_idx
s_feat_idx = src_node.feature_idx
# Grab decoders from local_model.clt for that feature
# For each layer in [s_layer..(node.layer_idx-1)], we can do something:
# out_vec = local_model.clt.W_dec[ index_map[s_layer][ℓ] ][ s_feat_idx ]
# partial = torch.dot( out_vec, grad_dict[ℓ, node.pos] ) # shape both [d_model]
# Summation of partial across the relevant layers.
sum_val = 0.0
# We'll define target_layer = node.layer_idx if node is feature
# or local_model.base.cfg.n_layers if node is logit
end_layer = (node.layer_idx if node_type == NodeType.FEATURE
else local_model.base.cfg.n_layers)
for mid_layer in range(s_layer, end_layer):
dec_idx = local_model.clt.index_map[s_layer][mid_layer]
out_vec = local_model.clt.W_dec[dec_idx][s_feat_idx] # shape [d_model]
# Suppose grad_dict[(mid_layer, node.context_pos)] is the gradient for that token/resid
# We'll do a dot product:
if (mid_layer, src_node.context_pos) not in grad_dict:
continue # might not exist
grad_vec = grad_dict[(mid_layer, src_node.context_pos)]
partial = torch.dot(out_vec, grad_vec)
sum_val += partial.item()
# Now multiply by a_s:
w = src_activation * sum_val
elif src_node.node_type == NodeType.EMBEDDING:
# Then the direct edge is basically
# w = (Emb_src)^T grad[ src_layer, context_pos ]
# Embeddings read into layer 0 (or wherever your model does).
# Or you might store them differently.
# If your local model lumps them into the "resid at layer 0," you can do:
emb_vec = src_node.embedding_vector
layer_for_emb = 0 # typically we treat the embedding as "layer 0"
pos = src_node.context_pos
if (layer_for_emb, pos) in grad_dict:
grad_vec = grad_dict[(layer_for_emb, pos)]
w = torch.dot(emb_vec, grad_vec).item()
else:
w = 0.0
elif src_node.node_type == NodeType.ERROR:
# Error nodes have no upstream inputs. By definition in the text,
# they "pop out of nowhere." So typically we do NOT add edges in from anything.
# Conversely, if you're computing edges from error to *this node*, you do:
# w = error_vec^T gradVec
# for the relevant layer. Example:
err_out_vec = src_node.output_vector # shape [d_model]
err_layer = src_node.layer_idx
pos = src_node.context_pos
if (err_layer, pos) in grad_dict:
grad_vec = grad_dict[(err_layer, pos)]
w = torch.dot(err_out_vec, grad_vec).item()
else:
w = 0.0
# If abs(w) is big enough, we add an edge
if abs(w) > 1e-2: # TODO: change this threshold to 1e-4
G.add_edge(src_id, node_id, w)
# done. G is updated with edges from source_node -> node_id.
# You'd repeat this function for each node in the graph for which
# you want to discover incoming edges.
return G # done
def build_attribution_graph_for_prompt(local_model, prompt_tokens):
# 1) build the nodes
G, cache = build_graph_nodes(prompt_tokens, local_model)
# 2) for each "target node" in logit_node_ids, do partial backward
for lnid in cache["logit_node_ids"]:
compute_direct_edges_for_node(G, lnid, local_model, prompt_tokens)
# 3) for each feature node, if you want edges going into it, do partial backward too
for (layer, pos, feat_i), node_id in cache["feature_nodes"].items():
compute_direct_edges_for_node(G, node_id, local_model, prompt_tokens)
# 4) done building adjacency
return G
def build_adjacency_matrix(G: AttributionGraph):
"""
Returns adjacency matrix A of shape [N, N], where A[j,i] = sum of edges from i->j
This is typically used for the pruning step.
"""
N = G.get_num_nodes()
A = np.zeros((N,N), dtype=np.float32)
for i in range(N):
for (j, w) in G.edges[i]:
A[j,i] += w
return A
def prune_graph(
G: AttributionGraph,
logit_nodes: List[int],
threshold_nodes=0.8,
threshold_edges=0.98
):
"""
1) Build adjacency matrix
2) compute B = (I - A)^-1 - I
3) For each node, sum up influence on logit nodes
4) keep top X% => subgraph
5) then re-build adjacency, do edge-level pruning similarly.
"""
# 1) adjacency
A = build_adjacency_matrix(G)
N = A.shape[0]
# 2) compute B
# we might want to clamp negative edges or do absolute value. The paper does "abs + row-normalize"
# For simplicity, do exactly as the paper says: we'll do a quick absolute + row normalization:
A_abs = np.abs(A)
row_sums = A_abs.sum(axis=1, keepdims=True)
row_sums[row_sums < 1e-10] = 1.0
A_norm = A_abs / row_sums
# Then B = (I - A_norm)^-1 - I
I = np.eye(N, dtype=np.float32)
M = (I - A_norm)
M_inv = np.linalg.inv(M)
B = M_inv - I # shape [N, N]
# 3) logit influence
# We have logit_nodes; we can weight them by their probability, or just sum them
# Suppose we do a simple sum
logit_mask = np.zeros((N,), dtype=np.float32)
for lnid in logit_nodes:
logit_mask[lnid] = 1.0
# node_influence = B * logit_mask
# shape = [N,N], [N], => we do a matmul
node_influence = B @ logit_mask # shape [N]
# 4) sort nodes by influence
order = np.argsort(-node_influence) # descending
csum = np.cumsum(node_influence[order])
total = csum[-1] if csum[-1]>0 else 1.0
cutoff_val = threshold_nodes*total
keep_mask = np.zeros((N,), dtype=bool)
for idx in range(len(order)):
if csum[idx] <= cutoff_val:
keep_mask[order[idx]] = True
else:
break
# We now form a new subgraph with only those nodes. (Edges from or to "kept" nodes.)
# Then we do a second pass for edge pruning using the same logic but with B for edges.
# For brevity, we'll just do node-level here.
# In practice, you'd want to do the 2-step approach from the text.
new_G = AttributionGraph()
old_to_new = {}
for i in range(N):
if keep_mask[i]:
new_i = new_G.add_node(G.nodes[i])
old_to_new[i] = new_i
for i in range(N):
if keep_mask[i]:
for (j, w) in G.edges[i]:
if keep_mask[j]:
new_G.add_edge(old_to_new[i], old_to_new[j], w)
return new_G
def compute_all_direct_edges_layer(
local_model,
tokens,
layer_idx,
target_nodes,
source_nodes,
threshold=1e-4,
freeze_ln=True,
freeze_attn=True,
):
"""
- Gathers the injection vectors for many 'target_nodes' that read from 'layer_idx'.
- Runs single_pass_grad_for_targets exactly ONCE, returning a 4D array of partial derivatives.
- For each source_node in `source_nodes`, does a dot product to get the direct edge
from source -> each target.
- Returns a list of (source_id, target_id, weight).
NOTE: This function is a simplified example. It only demonstrates:
- FEATURE nodes as targets, or LOGIT nodes as targets, for building injection vectors.
- EMBEDDING or FEATURE nodes as sources (we do a simple dot product).
- You can adapt it for your code (e.g. ERROR nodes, multi-layer partial sums, etc.).
"""
# 1) Build the injection matrix for the target_nodes
injection_list = []
# We'll keep track of which target_node corresponds to each row
# so we can output edges in the correct order
target_index_map = []
d_model = local_model.base.cfg.d_model
for i, tnode in enumerate(target_nodes):
if tnode.node_type == "feature":
# e.g. local_model.clt.W_enc[layer_idx][feat_idx]
feat_idx = tnode.feature_index
inj_vec = local_model.clt.W_enc[layer_idx][feat_idx].detach().clone()
elif tnode.node_type == "logit":
# logit injection
inj_vec = compute_logit_injection_vector(local_model, tnode.logit_index).detach().clone()
else:
# If you had embeddings or error nodes as targets, you'd define them here
raise ValueError(f"Unsupported target node type {tnode.node_type}")
injection_list.append(inj_vec)
target_index_map.append(i)
# shape = [n_targets, d_model]
injection_mat = torch.stack(injection_list, dim=0)
# 2) Single pass => gradient wrt this layer's residual
grad_wrt_resid = single_pass_grad_for_targets(
local_model=local_model,
tokens=tokens,
layer_idx=layer_idx,
target_injection_vectors=injection_mat,
freeze_ln=freeze_ln,
freeze_attn=freeze_attn,
)
# shape [batch, seq, d_model, n_targets]
# We'll assume batch=1 for simplicity
# (If batch>1, you might do a sum over all batch elements or handle them differently.)
grad_wrt_resid = grad_wrt_resid[0] # shape [seq, d_model, n_targets]
seq_len, d_model, n_targets = grad_wrt_resid.shape
# 3) Build edges from source_nodes to target_nodes
edges = [] # list of (src_id, tgt_id, weight)
for src_node in source_nodes:
src_id = src_node.id
if src_node.node_type == "embedding":
# Then the embedding vector is local_model.base.embed[idx], for token idx
# We'll do a dot product with grad_wrt_resid at the correct context_pos
emb_idx = src_node.logit_index # or however you store it
# Actually, we might store it as src_node.output_vector or something
# For demonstration:
token_id = src_node.logit_index
emb_vec = local_model.base.embed[token_id] # shape [d_model]
# Or if you stored an actual .embedding_vector in the node, use that
emb_vec = emb_vec.detach().clone()
pos = src_node.context_pos
if pos < 0 or pos >= seq_len:
# skip if out of range
continue
# shape [d_model, n_targets]
grad_slice = grad_wrt_resid[pos] # shape [d_model, n_targets]
# w = dot(emb_vec, grad_slice[:, i]) for each target i
# => shape [n_targets]
w_vals = torch.einsum("d, dn->n", emb_vec, grad_slice)
# w_vals is shape [n_targets]
# we can apply threshold, or store them all
for t_i, wval in enumerate(w_vals):
val = wval.item()
if abs(val) >= threshold:
tgt_id = target_nodes[t_i].id
edges.append((src_id, tgt_id, val))
elif src_node.node_type == "feature":
# Suppose we do a simpler, single-layer approach:
# direct edge ~ src_activation * dot(W_dec[src_feat], grad_slice)
# We'll find the correct position, assume it's src_node.context_pos
pos = src_node.context_pos
s_feat_idx = src_node.feature_index
# The src activation is typically stored in node.activation
src_activation = src_node.activation
# The "decoder" vector is local_model.clt.W_dec[index_map[src_layer][layer_idx]][s_feat_idx], etc.
# For simplicity, let's assume src_node.layer_idx == layer_idx
# in real code, you might do a loop from src_layer..layer_idx-1
if pos < 0 or pos >= seq_len:
continue
# shape [d_model, n_targets]
grad_slice = grad_wrt_resid[pos] # shape [d_model, n_targets]
# The decoder vector for that feature -> layer_idx
dec_idx = local_model.clt.index_map[src_node.layer_idx][layer_idx]
if dec_idx is None:
# skip if e.g. src_node.layer_idx>layer_idx
continue
out_vec = local_model.clt.W_dec[dec_idx][s_feat_idx] # shape [d_model]
# Then do dot(out_vec, grad_slice) => shape [n_targets]
partial_vals = torch.einsum("d, dn->n", out_vec, grad_slice)
# Multiply by the activation
w_vals = src_activation * partial_vals # shape [n_targets]
for t_i, wval in enumerate(w_vals):
val = wval.item()
if abs(val) >= threshold:
tgt_id = target_nodes[t_i].id
edges.append((src_id, tgt_id, val))
else:
# If you had other node types (ERROR, etc.), do similarly
pass
return edges
def build_direct_edges_single_pass_debug(
G: AttributionGraph,
local_model: LocalReplacementModel,
tokens: torch.Tensor,
threshold=1e-4,
):
"""
Debug version: We add print statements to confirm the shape of each injection vector
for each target node. We also print the final stacked shape, to confirm it's
[n_targets, d_model].
"""
n_layers = local_model.base.cfg.n_layers
d_model = local_model.base.cfg.d_model
print(f"[DEBUG] n_layers={n_layers}, d_model={d_model}")
# Bucket target nodes by layer
layer_to_targets = {L: [] for L in range(n_layers+1)}
for node in G.nodes:
if node.node_type == NodeType.LOGIT:
layer_to_targets[n_layers].append(node)
elif node.node_type == NodeType.FEATURE:
layer_to_targets[node.layer_idx].append(node)
else:
pass # e.g. embedding/error => sources only
# One pass per layer
for layer_i in range(n_layers+1):
target_nodes = layer_to_targets[layer_i]
if len(target_nodes) == 0:
continue
print(f"\n[DEBUG] Layer={layer_i}, #targets={len(target_nodes)}")
# 2A) Build injection_mat => shape [n_targets, d_model]
injection_list = []
for idx, tnode in enumerate(target_nodes):
if tnode.node_type == NodeType.FEATURE:
feat_idx = tnode.feature_index
w_enc_matrix = local_model.clt.W_enc[layer_i] # shape [num_features, d_model], e.g. [128,768]
print(f" [DEBUG] Target node {idx}: FEATURE layer={layer_i}, feat_idx={feat_idx}")
print(f" w_enc_matrix.shape = {tuple(w_enc_matrix.shape)}")
# The injection vector must be shape [d_model], not bigger
inj_vec = w_enc_matrix[feat_idx] # shape => [768]
print(f" inj_vec.shape = {tuple(inj_vec.shape)}")
injection_list.append(inj_vec.detach().clone())
elif tnode.node_type == NodeType.LOGIT:
print(f" [DEBUG] Target node {idx}: LOGIT layer={layer_i}, logit_index={tnode.logit_index}")
inj_vec = compute_logit_injection_vector(local_model, tnode.logit_index)
print(f" inj_vec.shape = {tuple(inj_vec.shape)}")
injection_list.append(inj_vec.detach().clone())
else:
print(f" [DEBUG] Skipping target node {idx}: node_type={tnode.node_type}")
continue
if len(injection_list) == 0:
print(f" [DEBUG] No valid feature/logit targets for layer={layer_i}, skipping.")
continue
# shape => [n_targets, d_model]
injection_mat = torch.stack(injection_list, dim=0)
print(f"[DEBUG] injection_mat final shape => {tuple(injection_mat.shape)}")
# If layer_i == n_layers => clamp to n_layers-1
actual_layer = min(layer_i, n_layers - 1)
print(f"[DEBUG] actual_layer = {actual_layer}")
# 2B) single backward pass => gradient wrt resid at actual_layer
grad_wrt_resid = single_pass_grad_for_targets_debug(
local_model=local_model,
tokens=tokens,
layer_idx=actual_layer,
target_injection_vectors=injection_mat,
freeze_ln=True,
freeze_attn=True,
)
# => shape [batch=1, seq, d_model, n_targets]
grad_wrt_resid = grad_wrt_resid[0] # => [seq, d_model, n_targets]
seq_len, _, n_targets = grad_wrt_resid.shape
# 2C) For each source node...
for src_id, src_node in enumerate(G.nodes):
if not is_upstream_layer_layer(src_node, layer_i):
continue
for t_idx, tgt_node in enumerate(target_nodes):
pos = tgt_node.context_pos
if (pos < 0) or (pos >= seq_len):
continue
grad_slice = grad_wrt_resid[pos, :, t_idx] # => shape [d_model]
w_val = 0.0
if src_node.node_type == NodeType.EMBEDDING:
emb_vec = src_node.output_vector # shape [d_model]
w_val = float(torch.dot(emb_vec, grad_slice))
elif src_node.node_type == NodeType.FEATURE:
a_s = src_node.activation
s_feat_idx = src_node.feature_index
s_layer = src_node.layer_idx
target_index = min(layer_i, n_layers - 1)
dec_idx = local_model.clt.index_map[s_layer][target_index]
if dec_idx is not None:
out_vec = local_model.clt.W_dec[dec_idx][s_feat_idx] # [d_model]
partial = float(torch.dot(out_vec, grad_slice))
w_val = a_s * partial
if abs(w_val) >= threshold:
G.add_edge(src_id, tgt_node.id, w_val)
return G
def single_pass_grad_for_targets_debug(
local_model,
tokens,
layer_idx,
target_injection_vectors,
freeze_ln=True,
freeze_attn=True,
):
"""
Same as single_pass_grad_for_targets but with debug prints about shapes.
"""
d_model = local_model.base.cfg.d_model
print(f"\n[DEBUG single_pass_grad_for_targets] => layer_idx={layer_idx}")
print(f" target_injection_vectors.shape = {list(target_injection_vectors.shape)}")
local_model.zero_grad(set_to_none=True)
# Optionally freeze LN or QK
if freeze_ln or freeze_attn:
for name, param in local_model.base.named_parameters():
if freeze_ln and ("ln" in name):
param.requires_grad_(False)
if freeze_attn and (".W_Q" in name or ".W_K" in name):
param.requires_grad_(False)
# Hook
storage = {}
def store_resid_hook(resid, hook):
storage["resid"] = resid
return resid
# E.g. blocks.{layer_idx}.hook_resid_mid
resid_name = f"blocks.{layer_idx}.hook_resid_mid"
if resid_name not in local_model.base.hook_dict:
# fallback
resid_name = f"blocks.{layer_idx}.hook_resid_post"
handle = local_model.base.add_hook(resid_name, store_resid_hook, "fwd")
with torch.enable_grad():
_ = local_model(tokens)
if handle is not None:
handle.remove()
if "resid" not in storage:
raise ValueError(f"Could not find residual for layer {layer_idx}: {resid_name}")
resid_L = storage["resid"]
print(f" resid_L.shape = {tuple(resid_L.shape)}")
# Now we verify the injection_vec shape is [n_targets, d_model].
n_targets = target_injection_vectors.shape[0]
if target_injection_vectors.ndim != 2 or target_injection_vectors.shape[1] != d_model:
raise RuntimeError(
f"[DEBUG ERROR] Expected injection_vectors shape [n_targets, {d_model}], "
f"but got {list(target_injection_vectors.shape)}"
)
# 4) build partial losses
injection_mat = target_injection_vectors.view(1, 1, d_model, n_targets)
expanded_resid = resid_L.unsqueeze(-1) # => [batch, seq, d_model, 1]
product = expanded_resid * injection_mat # => [batch, seq, d_model, n_targets]
partial_losses = product.sum(dim=[0,1,2]) # => shape [n_targets]
# 3) Provide grad_outputs => identity, so we get full Jacobian
outs = partial_losses # shape [n_targets]
jac_list = []
for i in range(n_targets):
unit_vec = torch.zeros_like(outs) # shape [n_targets]
unit_vec[i] = 1.0
# Now partial derivative of outs·unit_vec => outs[i]
grad_i = torch.autograd.grad(
outputs=outs, # [n_targets]
inputs=resid_L, # [batch, seq, d_model]
grad_outputs=unit_vec, # also [n_targets]
retain_graph=True,
)[0] # => shape [batch, seq, d_model]
jac_list.append(grad_i.unsqueeze(-1)) # => [batch, seq, d_model, 1]
print(f"index={i}, grad_i.shape={tuple(grad_i.shape)}")
# Finally cat them => [batch, seq, d_model, n_targets]
grad_wrt_resid = torch.cat(jac_list, dim=-1)
print(f" => grad_wrt_resid.shape = {tuple(grad_wrt_resid.shape)}\n")
return grad_wrt_resid
putting it together so far
#### collect 5k samples
main_collect_5k()
#### train the CLT
dataset = CLTHiddenStateDataset("clt_data")
dataloader = DataLoader(dataset, batch_size=1, shuffle=True, num_workers=4)
model_name = "gpt2-small"
hmodel = HookedTransformer.from_pretrained(model_name)
layer_feature_list = [128]*hmodel.cfg.n_layers # or something custom
cross_layer_transcoder = CrossLayerTranscoder.from_hookedtransformer(
hmodel, layer_feature_list, bandwidth=1.0, device="cuda"
)
train_clt(cross_layer_transcoder, dataloader, 2)
#### build the local replacement model
prompt = "Hello, I'd like you to help me plan my wedding please."
local_replacement_model = build_local_replacement_model_with_cache(hmodel, cross_layer_transcoder, prompt, device="cuda")
#### Let's compare outputs of the original model vs. the local replacement
with torch.no_grad():
tokens = hmodel.to_tokens(prompt, prepend_bos=True).to("cuda")
orig_logits = hmodel(tokens)
rep_logits = local_replacement_model(tokens)
print("Are the logits close?", torch.allclose(orig_logits, rep_logits, atol=1e-5))
tokens = hmodel.to_tokens(prompt, prepend_bos=True)
#### 1) Build node set
G, cache = build_graph_nodes(local_replacement_model, prompt, top_k=3)
#### 2) compute direct edges for each logit node or MLP node
build_direct_edges_single_pass_debug(G, local_replacement_model, tokens, threshold=1e-4)
#### 3) prune
pruned_G = prune_graph(G, cache["logit_node_ids"],
threshold_nodes=0.8,
threshold_edges=0.98)
there ya go. some breakdown of anthropic’s paper with the assistance of some of my dearest friends: sonnet 3.7 thinking, o1-pro, gpt-4o, o3-mini-high, and long time homie, sonnet 3.5.