Why So Hard (Negative) On Your Self (Reinforcement)?
==================
Intro
My main goal here was simple. I just want to see the impact of hard-negative choices on a modern embedder. Nothing more, nothing less.
I noticed a model trained with BM25 hard-negatives performed extremely strongly and I wanted to ask the question ‘why?’. For a bit of background, Filevine is a legal tech company where I currently work where I’m inundated with legal text of every variety. What’s striking about the legal text is that it can range from nonsensical to extremely logical and is almost always set up like some kind of discrete math word problem. It’s very funky stuff. My gut hypothesis is this is probably why most frontier models suck at law right now - is because it appears normal, it’s not super varied (so i think a lot gets filtered out in pretraining) - and most of the juicy stuff isn’t public.
Anyway, back to the show: We’ll take a standard dataset (legal summarization; another constraint) find hard-negatives via bm25, find negatives through a high-ranked (on MTEB) embedding model (Alibaba-NLP/gte-Qwen2-1.5B-instruct) + high ranked re-ranker (Alibaba-NLP/gte-multilingual-reranker-base), and lastly the high-ranked embedding model alone (Alibaba-NLP/gte-multilingual-reranker-base). We’ll do this to see the effect on hard-negative choice and see how well the choices impact downstream legal retrieval tasks
Experimental Setup
We’re going to take the dataset of joelniklaus/legal_case_document_summarization (selected after eyeballing the data and gut feeling that it ‘looked good’ – highly scientific, I know) and use that to create a legal training dataset and then test it on 6 MTEB legal tasks: LegalBenchCorporateLobbying, LegalSummarization, AILACasedocs, LegalQuAD, LegalBenchConsumerContractsQA, AILAStatutes.
some notes on the evals:
AILA tasks:
These tasks require some kind of knowledge about Indian law (which we don’t have).
Task 1: Identifying relevant prior cases
We provide ~3000 case documents of cases that were judged in the Supreme Court of India. For each query, the task is to retrieve the most similar / relevant case document with respect to the situation in the given query.
Task 2: Identifying relevant statutes
We have identified a set of 197 statutes (Sections of Acts) from Indian law, that are relevant to some of the queries. We provide the title and description of these statutes. For each query, the task is to identify the most relevant statutes (from among the 197 statutes). Note that, the task can be modelled either as an unsupervised retrieval task (where you search for relevant statues) or as a supervised classification task (e.g., trying to predict for each statute whether it is relevant). For the latter, case documents provided for Task 1 can be utilized. However, if a team wishes to apply supervised models, then it is their responsibility to create the necessary training data.
LegalQuAD
This is a German dataset - and our training set is not german - so duh, I wouldn’t expect it to be useful for us given that we won’t have any german contrastive pairs, but we’ll throw it in there to see what kind of random effects we can see.
Basic Analysis:
I want to do some basic analysis to understand this legal dataset by comparing it against other domain-specific (math, medicine) and general data (openweb) to showcase law data to the reader
Part of Speech Distribution
def get_pos_distribution(documents, sample_size=1000):
"""
For efficiency, you may want to sample some documents
if the dataset is very large.
"""
sampled_docs = documents[:sample_size]
pos_counts = Counter()
total_tokens = 0
for doc_text in sampled_docs:
doc = nlp(doc_text)
for token in doc:
pos_counts[token.pos_] += 1
total_tokens += 1
# Convert counts to proportions
pos_dist = {pos: count / total_tokens for pos, count in pos_counts.items()}
return pos_dist
pos_legal = get_pos_distribution(legal_docs['judgement'])
pos_math = get_pos_distribution(math_docs['prompt'])
pos_openweb = get_pos_distribution(openweb_docs['text'])
pos_math2= get_pos_distribution(other_math['question'])
pos_med = get_pos_distribution(med['page_text'])
# Convert to DataFrame for easy comparison
df_pos = pd.DataFrame([pos_legal, pos_math, pos_openweb, pos_math2, pos_med],
index=["Legal", "Math", "OpenWeb", "Word Math", "Wiki Med"]).fillna(0)
df_pos = df_pos.transpose().sort_values(by=["Legal", "Math", "OpenWeb", "Word Math", "Wiki Med"], ascending=False)
print(df_pos)
Legal Math OpenWeb Word Math Wiki Med
NOUN 0.175572 0.189513 0.175243 0.210162 0.249462
ADP 0.135626 0.068894 0.097755 0.097116 0.112305
DET 0.125641 0.071518 0.079878 0.091713 0.071084
VERB 0.090413 0.053803 0.105108 0.084351 0.087313
PUNCT 0.089545 0.167888 0.125426 0.152996 0.122003
PROPN 0.076987 0.064055 0.087082 0.039782 0.050926
AUX 0.060485 0.036361 0.048429 0.048208 0.050527
PRON 0.042584 0.030702 0.064171 0.042777 0.020965
ADJ 0.041660 0.049893 0.061888 0.057811 0.102179
NUM 0.029754 0.077861 0.020058 0.088942 0.017541
CCONJ 0.029093 0.025234 0.028108 0.032279 0.034960
SPACE 0.026921 0.032041 0.020384 0.000056 0.021435
SCONJ 0.025912 0.015993 0.018093 0.026596 0.011554
PART 0.023696 0.004484 0.025851 0.012094 0.012989
ADV 0.023398 0.009787 0.036608 0.013690 0.032312
X 0.002462 0.019848 0.002509 0.001092 0.000881
SYM 0.000129 0.081552 0.002469 0.000280 0.001497
INTJ 0.000122 0.000574 0.000937 0.000056 0.000067
Named Entity Recogniton Distributin
def get_ner_distribution(documents, sample_size=1000):
sampled_docs = documents[:sample_size]
ner_counts = Counter()
total_ents = 0
for doc_text in sampled_docs:
doc = nlp(doc_text)
for ent in doc.ents:
ner_counts[ent.label_] += 1
total_ents += 1
ner_dist = {ent_type: count / total_ents for ent_type, count in ner_counts.items()} if total_ents > 0 else {}
return ner_dist
ner_legal = get_ner_distribution(legal_docs['judgement'])
ner_math = get_ner_distribution(math_docs['prompt'])
ner_openweb = get_ner_distribution(openweb_docs['text'])
ner_math2 = get_ner_distribution(other_math['question'])
ner_med = get_ner_distribution(med['page_text'])
df_ner = pd.DataFrame([ner_legal, ner_math, ner_openweb, ner_math2, ner_med],
index=["Legal", "Math", "OpenWeb", "Word Math", "Wiki Med"]).fillna(0)
df_ner = df_ner.transpose().sort_values(by=["Legal", "Math", "OpenWeb", "Word Math", "Wiki Med"], ascending=False)
print(df_ner)
Legal Math OpenWeb Word Math Wiki Med
ORG 0.263405 0.068710 0.213852 0.040993 0.253864
CARDINAL 0.238252 0.252273 0.118554 0.517965 0.164357
DATE 0.131707 0.081172 0.138075 0.095008 0.153279
PERSON 0.126937 0.181543 0.218140 0.083193 0.112978
LAW 0.104598 0.002358 0.003750 0.000723 0.000867
GPE 0.053235 0.017178 0.121413 0.048469 0.107683
ORDINAL 0.027613 0.031324 0.022764 0.034242 0.027509
NORP 0.016257 0.008420 0.060991 0.011575 0.051120
WORK_OF_ART 0.008050 0.000337 0.013449 0.000723 0.006770
LOC 0.007295 0.001347 0.012527 0.000482 0.016860
PRODUCT 0.007116 0.008420 0.009745 0.005787 0.011197
FAC 0.004185 0.001010 0.009714 0.000482 0.002543
TIME 0.003254 0.019198 0.014541 0.016639 0.012374
QUANTITY 0.002291 0.022903 0.007363 0.139860 0.008804
MONEY 0.002089 0.301785 0.016201 0.000241 0.003460
EVENT 0.001604 0.000337 0.006563 0.000482 0.003500
PERCENT 0.001207 0.001684 0.009929 0.000241 0.061709
LANGUAGE 0.000905 0.000000 0.002429 0.002894 0.001127
BERT Embeddings Clusters
model = SentenceTransformer('all-MiniLM-L6-v2').cpu()
def get_embeddings(documents, sample_size=10000):
sample_docs = documents[:sample_size]
embeddings = model.encode(sample_docs, show_progress_bar=True)
return embeddings
emb_legal = get_embeddings(legal_docs['judgement'])
emb_math = get_embeddings(math_docs['prompt'])
emb_openweb = get_embeddings(openweb_docs['text'])
emb_other_math = get_embeddings(other_math['question'])
emb_med = get_embeddings(med['page_text'])
# Combine them with labels
all_embeddings = np.vstack([emb_legal, emb_math, emb_openweb, emb_other_math, emb_med])
labels = (["Legal"] * len(emb_legal)) + (["Math"] * len(emb_math)) + (["OpenWeb"] * len(emb_openweb)) + (["Orca Math Word Problems"] * len(emb_other_math)) + (["Wiki Med"] * len(emb_med))
# Apply UMAP or t-SNE
reducer = umap.UMAP(n_neighbors=15, min_dist=0.1, random_state=42)
# Alternatively: reducer = TSNE(n_components=2, random_state=42)
embedding_2d = reducer.fit_transform(all_embeddings)
# Plot
plt.figure(figsize=(10, 8))
sns.scatterplot(
x=embedding_2d[:,0],
y=embedding_2d[:,1],
hue=labels,
palette="deep"
)
plt.title("Sentence-BERT Embeddings (UMAP) - Dataset Clusters")
plt.show()
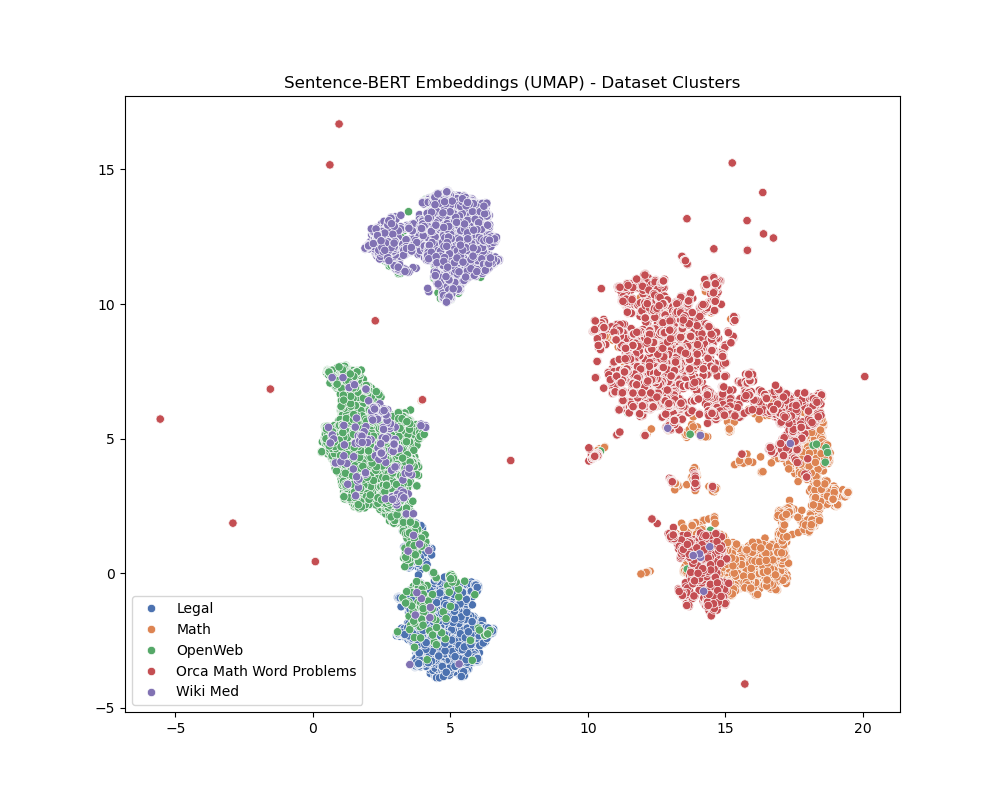
Med and Math are on their own islands while Legal shares a lot of similarities with ‘regular text’. And in my experience, this checks - it uses ‘normal words’ but uses them differently.
Lexical Entropy
import math
from collections import Counter
import nltk
from nltk.tokenize import word_tokenize
nltk.download('punkt')
def compute_lexical_entropy(documents):
"""
Computes the lexical entropy (in bits) for a list of documents.
H(X) = - sum( p(x) * log2(p(x)) ) for all x in the vocabulary.
"""
freq = Counter()
from nltk.corpus import stopwords
stop_words = set(stopwords.words('english'))
for doc in documents:
word_tokens = word_tokenize(doc.lower())
word_tokens = [re.sub(r'[^\w\s]', '', token) for token in word_tokens if re.sub(r'[^\w\s]', '', token)]
word_tokens_counter = Counter(word_tokens)
freq.update(word_tokens_counter)
total_tokens = sum(freq.values())
# Calculate entropy
entropy = 0.0
for word, count in freq.items():
p = count / total_tokens
entropy -= p * math.log2(p) # minus sign because p*log2(p) is negative
return entropy
entropy_legal = compute_lexical_entropy(legal_docs['judgement'])
entropy_math = compute_lexical_entropy(math_docs['prompt'])
entropy_openweb = compute_lexical_entropy(openweb_docs['text'])
entropy_other_math = compute_lexical_entropy(other_math['question'])
entropy_med = compute_lexical_entropy(med['page_text'])
print(f"Lexical Entropy (Legal Summarization): {entropy_legal:.4f} bits")
print(f"Lexical Entropy (Competition Math): {entropy_math:.4f} bits")
print(f"Lexical Entropy (OpenWeb): {entropy_openweb:.4f} bits")
print(f"Lexical Entropy (Math Word Problems): {entropy_other_math:.4f} bits")
print(f"Lexical Entropy (Wiki Med): {entropy_med:.4f} bits")
Lexical Entropy (Legal Summarization): 9.5891 bits
Lexical Entropy (Competition Math): 9.1562 bits
Lexical Entropy (OpenWeb): 11.0669 bits
Lexical Entropy (Math Word Problems): 9.4199 bits
Lexical Entropy (Wiki Med): 10.9722 bits
OpenWeb has the highest lexical entropy (~11.07), suggesting the broadest or most diverse vocabulary and less repetition of the same tokens. Wiki Med is second highest (10.97), indicating it also uses a fairly large and varied vocabulary—likely due to the breadth of medical topics while Legal Summarization sits in the middle (~9.59), higher than the math corpora but lower than OpenWeb and Wiki Med.
The Math datasets (Competition Math and Math Word Problems) have the lowest entropy (~9.16–9.42), consistent with a narrower or more specialized vocabulary (repetitions of numbers, math symbols, certain keywords).
Higher entropy typically means more linguistic variety and the math datasets are more repetitive or domain-specific (numbers, symbols, certain repeated words). Legal is “in-between” with some specialized vocabulary, but it’s not as wide-ranging as something like OpenWeb or Wiki Med, which cover many topics.
Readability
import statistics
import textstat
from nltk.tokenize import sent_tokenize
def compute_readability_scores(documents, sample_size=1000):
"""
Computes average readability scores on a subset of the dataset
to avoid extremely large computations.
Returns: (avg_flesch, avg_fog)
"""
# Sample to speed up (adjust as needed)
sampled_docs = documents[:sample_size]
flesch_scores = []
fog_scores = []
for doc in sampled_docs:
# textstat expects raw text (sentences, words)
flesch = textstat.flesch_reading_ease(doc)
fog = textstat.gunning_fog(doc)
flesch_scores.append(flesch)
fog_scores.append(fog)
avg_flesch = statistics.mean(flesch_scores)
avg_fog = statistics.mean(fog_scores)
return avg_flesch, avg_fog
# Example usage
legal_flesch, legal_fog = compute_readability_scores(legal_docs['judgement'])
math_flesch, math_fog = compute_readability_scores(math_docs['prompt'])
openweb_flesch, openweb_fog = compute_readability_scores(openweb_docs['text'])
other_math_flesch, other_math_fog = compute_readability_scores(other_math['question'])
med_flesch, med_fog = compute_readability_scores(med['page_text'])
print("==== Flesch Reading Ease (Higher = Easier to Read) ====")
print(f"Legal Summarization: {legal_flesch:.2f}")
print(f"Competition Math: {math_flesch:.2f}")
print(f"OpenWeb: {openweb_flesch:.2f}")
print(f"Math Word Problems: {other_math_flesch:.2f}")
print(f"Wiki Med: {med_flesch:.2f}")
print("\n==== Gunning Fog Index (Lower = Easier to Read) ====")
print(f"Legal Summarization: {legal_fog:.2f}")
print(f"Competition Math: {math_fog:.2f}")
print(f"OpenWeb: {openweb_fog:.2f}")
print(f"Math Word Problems: {other_math_fog:.2f}")
print(f"Wiki Med: {med_fog:.2f}")
==== Flesch Reading Ease (Higher = Easier to Read) ====
Legal Summarization: 54.08
Competition Math: 75.10
OpenWeb: 59.90
Math Word Problems: 82.43
Wiki Med: 36.06
==== Gunning Fog Index (Lower = Easier to Read) ====
Legal Summarization: 12.70
Competition Math: 7.85
OpenWeb: 11.40
Math Word Problems: 7.45
Wiki Med: 13.38
High Flesch, Low Fog = simpler sentences, shorter words, more straightforward syntax. Low Flesch, High Fog = complex sentences, longer words, domain-specific jargon
Wiki Med is the hardest to read, eg the most domain-heavy in vocabulary and syntax. Legal Summarization is also formal, but not as severe as Wiki Med. The math text is ‘easy’ (“Find x if …,” “What is the value of …,” etc.) - but given that we know it’s technically ‘hard’, this sort of metric doesn’t really tell us anything.
Quick Conclusions:
OpenWeb = high diversity, moderate difficulty.
Wiki Med = high diversity, highest difficulty (lots of specialized medical terms).
Legal Summarization = medium diversity, high difficulty (formal style).
Math Word Problems & Competition Math = lower diversity, easier style (short sentences, specialized numeric vocab, but not big words).
I don’t know how to visualize my bias yet that legal text is like discrete math word problems, vocalized in specialized text (like medical) that are extremely long. Just having gone through many legal docs of late, that’s how I’ve come to think about it. I think it gives the appearance of having overlap with webtext, but it’s ‘just not’. And I think this largely comes from it being a protocol/communication layer over morals/values/ethics where it uses the framework/logic to construct text that “isn’t strictly logical” (sometimes). “1 + 1 = 3 because ‘jury’ and here’s 10000 words on why.”
It’s very hard and I really don’t think many models so far have been able to grok it. Onward.
Code
In this portion, I’ll provide some of the code I used for generating the initial hard negatives before this morphed into my own personal rabbit-hole project.
# Standard library
import hashlib
from typing import List, Dict, Any, Tuple, Optional, Union
from collections import deque
from concurrent.futures import ThreadPoolExecutor
# Deep learning & numerical
import torch
import numpy as np
import faiss
from sentence_transformers import SentenceTransformer
from transformers import AutoModelForSequenceClassification, AutoTokenizer
# Data handling
from datasets import Dataset, load_dataset, load_from_disk
# Text processing
import bm25s
import Stemmer # For stemming
import pickle
class FastEmbedder(mp.Process):
"""Parallel processor for creating document embeddings using GPU acceleration."""
def __init__(
self,
gpu_id: int,
input_queue: mp.Queue,
output_queue: mp.Queue,
model_id: str = "Alibaba-NLP/gte-Qwen2-1.5B-instruct",
batch_size: int = 256,
embedding_size: int = 1536
):
"""Initialize the FastEmbedder process.
Args:
gpu_id: GPU device identifier
input_queue: Queue for receiving document batches
output_queue: Queue for sending computed embeddings
model_id: Identifier for the embedding model
batch_size: Number of documents to process at once
embedding_size: Dimension of the embedding vectors
"""
super().__init__()
self.gpu_id = gpu_id
self.input_queue = input_queue
self.output_queue = output_queue
self.model_id = model_id
self.batch_size = batch_size
self.embedding_size = embedding_size
def setup(self):
"""Initialize GPU device and load model."""
self.device = torch.device(f"cuda:{self.gpu_id}")
torch.cuda.set_device(self.device)
self.model = SentenceTransformer(self.model_id).cuda()
self.model.half()
def process_batch(self, batch: Dict[str, Any], current_batch_size: int, max_seq_len: int = 32768) -> Dict[str, Any]:
"""Process a batch of documents and create embeddings.
Args:
batch: Dictionary containing texts and their indices
current_batch_size: Current batch size for processing
max_seq_len: Maximum sequence length for tokenization
Returns:
Dictionary containing embeddings and document indices
"""
start_idx = 0
all_embeddings = []
all_indices = []
while start_idx < len(batch['texts']):
end_idx = start_idx + current_batch_size
try:
doc_texts = batch['texts'][start_idx:end_idx]
doc_indices = batch['doc_indices'][start_idx:end_idx]
tokenized = self.model.tokenizer(
doc_texts,
truncation=True,
max_length=max_seq_len
)
truncated_texts = self.model.tokenizer.batch_decode(
tokenized['input_ids'],
skip_special_tokens=True
)
embeddings = self.model.encode(
truncated_texts,
convert_to_numpy=True,
normalize_embeddings=True
).astype(np.float32)
torch.cuda.empty_cache()
all_embeddings.extend(embeddings)
all_indices.extend(doc_indices)
start_idx = end_idx
except RuntimeError as e:
if "CUBLAS_STATUS_ALLOC_FAILED" in str(e):
self.model.to('cpu')
torch.cuda.synchronize(self.gpu_id)
torch.cuda.empty_cache()
torch.cuda.set_device(self.gpu_id)
self.setup()
return self.process_batch(batch, current_batch_size, max_seq_len)
if "out of memory" in str(e):
torch.cuda.empty_cache()
if max_seq_len > 1024:
new_seq_len = max_seq_len // 2
return self.process_batch(batch, current_batch_size, new_seq_len)
self.batch_size = max(1, current_batch_size // 2)
return self.process_batch(batch, self.batch_size, max_seq_len)
raise e
return {
'embeddings': np.array(all_embeddings),
'doc_indices': all_indices
}
def run(self):
"""Main process loop for embedding generation."""
self.setup()
while True:
batch = self.input_queue.get()
if batch is None:
self.output_queue.put(None)
break
result = self.process_batch(batch, self.batch_size)
self.output_queue.put(result)
class ParallelFaissSearcher(mp.Process):
"""Parallel processor for finding hard negatives using FAISS similarity search."""
def __init__(
self,
gpu_id: int,
input_queue: mp.Queue,
output_queue: mp.Queue,
index: faiss.IndexFlatIP,
faiss_to_doc_idx: Dict[int, int],
doc_list: List[str],
query_idx_relations: Dict[int, Dict[str, set]],
rerank: bool = True,
model_id: str = "cross-encoder/ms-marco-MiniLM-L-6-v2",
faiss_use_gpu: bool = True
):
"""Initialize the ParallelFaissSearcher process.
Args:
gpu_id: GPU device identifier
input_queue: Queue for receiving query batches
output_queue: Queue for sending results
index: FAISS index for similarity search
faiss_to_doc_idx: Mapping from FAISS indices to document indices
doc_list: List of original documents
query_idx_relations: Mapping of query relationships
rerank: Whether to use reranking
model_id: Identifier for reranking model
faiss_use_gpu: Whether to use GPU for FAISS
"""
super().__init__()
self.gpu_id = gpu_id
self.input_queue = input_queue
self.output_queue = output_queue
self.cpu_index = index
self.faiss_to_doc_idx = faiss_to_doc_idx
self.doc_list = doc_list
self.query_idx_relations = query_idx_relations
self.rerank = rerank
self.model_id = model_id
self.faiss_use_gpu = faiss_use_gpu
def setup(self):
"""Initialize GPU resources and models."""
if self.faiss_use_gpu:
res = faiss.StandardGpuResources()
# load here
cpu_index = faiss.read_index(self.cpu_index)
self.index = faiss.index_cpu_to_gpu(res, self.gpu_id, cpu_index)
print(f"FAISS success on gpu: {self.gpu_id}")
else:
self.index = faiss.read_index(self.cpu_index)
if self.rerank:
self.tokenizer = AutoTokenizer.from_pretrained(self.model_id)
self.reranker = AutoModelForSequenceClassification.from_pretrained(
self.model_id, trust_remote_code=True,
torch_dtype=torch.float16
).to(f"cuda:{self.gpu_id}")
self.reranker.eval()
def find_hard_negative(self, query_idx: int, k: int = 10) -> Optional[int]:
"""Find hard negative example for a given query.
Args:
query_idx: Index of the query document
k: Number of nearest neighbors to consider
Returns:
Index of the selected hard negative, or None if not found
"""
print(f"gpu: {self.gpu_id}: {query_idx}")
query_text = self.doc_list[query_idx]
positive_idxs = self.query_idx_relations[query_idx]['positive']
faiss_query_idx = [idx for idx, doc_idx in self.faiss_to_doc_idx.items()
if doc_idx == query_idx][0]
query_embedding = self.index.reconstruct(faiss_query_idx).reshape(1, -1)
print(f"gpu: {self.gpu_id}: reconstructing {query_idx}")
D, I = self.index.search(query_embedding, k)
print(f"gpu: {self.gpu_id}: search-passed {query_idx}")
candidates = []
for faiss_idx in I[0]:
doc_idx = self.faiss_to_doc_idx[faiss_idx]
if doc_idx != query_idx and doc_idx not in positive_idxs:
candidates.append((doc_idx, self.doc_list[doc_idx]))
if not self.rerank:
return candidates[0][0] if candidates else None
if candidates:
pairs = [[query_text, c[1]] for c in candidates]
with torch.no_grad():
inputs = self.tokenizer(pairs, padding=True, truncation=True, return_tensors='pt', max_length=32768)
inputs = {k: v.to(f"cuda:{self.gpu_id}") for k, v in inputs.items()}
scores = self.reranker(**inputs, return_dict=True).logits.view(-1, ).float()
# print(scores)
# scores = self.reranker.predict(pairs)
best_idx = max(range(len(scores)), key=lambda i: scores[i])
return candidates[best_idx][0]
return None
def run(self):
"""Main process loop for finding hard negatives."""
self.setup()
while True:
batch = self.input_queue.get()
if batch is None:
self.output_queue.put(None)
break
results = {}
for query_idx in batch:
result = self.find_hard_negative(query_idx)
if result:
results[query_idx] = result
self.output_queue.put(results)
def create_bm25_data(dataset_config: dict, save_name: str = 'summarizer_bm25') -> Dataset:
"""Create training dataset using BM25 retrieval for hard negative mining.
This function processes legal documents and summaries to create training examples
with hard negative pairs selected using BM25 similarity scores.
Args:
save_name: Name for saving the processed dataset
Returns:
Dataset containing query-positive-negative triplets
"""
def get_hard_negatives(
query: str,
positive_ids: set,
stemmer,
retriever,
k: int = 2,
threshold: float = 0.5,
single: bool = True
) -> Union[Tuple[str, float], List[Tuple[str, float]]]:
"""Get hard negative examples that are semantically similar but not relevant.
Args:
query: Input text to find negatives for
positive_ids: Set of known positive document IDs
stemmer: Stemmer for text preprocessing
retriever: BM25 retriever instance
k: Number of negatives to retrieve
threshold: Similarity threshold for hard negatives
single: Whether to return single negative or multiple
Returns:
Either tuple of (negative_text, score) or list of such tuples
"""
query_tokens = bm25s.tokenize(query, stemmer=stemmer)
results, scores = retriever.retrieve(query_tokens, k=len(corpus))
if single:
for i in range(results.shape[1]):
doc_id, score = results[0, i], scores[0, i]
if 0 < score < threshold and doc_id not in positive_ids:
return corpus[doc_id], score
return None, 0.0
hard_negatives = []
for i in range(results.shape[1]):
doc_id, score = results[0, i], scores[0, i]
if 0 < score < threshold and doc_id not in positive_ids:
hard_negatives.append((corpus[doc_id], score))
if len(hard_negatives) >= k:
break
return hard_negatives
def process_query_threaded(query_data: Tuple[str, List[str]], shared_stemmer, shared_retriever) -> List[Tuple[str, str, Tuple[str, float]]]:
"""Process a single query to generate training examples using shared resources.
Args:
query_data: Tuple of (query_text, positive_examples)
shared_stemmer: Stemmer instance shared across threads
shared_retriever: BM25 retriever shared across threads
Returns:
List of (query, positive, negative) training examples
"""
query, positives = query_data
positives = deque(positives)
len_positives = len(positives)
positives = [positives] if isinstance(positives, str) else positives
positive_ids = {i for i, doc in enumerate(corpus) if doc in positives}
results = []
while positives:
pos = positives.popleft()
neg = get_hard_negatives(
query,
positive_ids,
shared_stemmer,
shared_retriever,
k = 500 if len_positives < 10 else len_positives+500,
threshold = 0.5,
single=True
)
if neg:
results.append((query, pos, neg))
return results
# Initialize data structures
train_data = {
"query": [],
"positive": [],
"negative": []
}
queries = {}
ds = load_dataset(dataset_config["name"])
for split in dataset_config["splits"]:
for text1_col, text2_col in dataset_config["pairs"]:
for item in ds[split]:
text1, text2 = item[text1_col], item[text2_col]
for base_text, paired_text in [(text1, text2), (text2, text1)]:
if base_text not in queries:
queries[base_text] = {"positive": [], "negative": []}
queries[base_text]["positive"].append(paired_text)
# Create BM25 index
corpus = list(queries.keys())
stemmer = Stemmer.Stemmer("english")
corpus_tokens = bm25s.tokenize(corpus, stopwords="en", stemmer=stemmer)
retriever = bm25s.BM25()
retriever.index(corpus_tokens)
# Process queries in parallel
with ThreadPoolExecutor(max_workers=96) as executor:
query_data = [(query, queries[query]['positive']) for query in queries]
futures = [executor.submit(process_query_threaded, qd, stemmer, retriever) for qd in query_data]
for future in futures:
for query, pos, (neg, score) in future.result():
if neg:
train_data['query'].append(query)
train_data['positive'].append(pos)
train_data['negative'].append(neg)
# Create and filter dataset
dataset = Dataset.from_dict(train_data)
valid_indices = [i for i in range(len(dataset))
if not any(dataset[i][field] is None
for field in ['query', 'positive', 'negative'])]
dataset = dataset.select(valid_indices)
dataset.save_to_disk(save_name)
return dataset
def create_fast_index(
doc_list: List[str],
num_gpus: int = 8,
batch_size: int = 256,
embedding_size: int = 1536,
model_id: str = "Alibaba-NLP/gte-Qwen2-1.5B-instruct"
) -> Tuple[faiss.IndexFlatIP, Dict[int, int]]:
"""Create FAISS index from document list using parallel processing.
Args:
doc_list: List of documents to embed
num_gpus: Number of GPUs to use
batch_size: Batch size per GPU
embedding_size: Dimension of embeddings
model_id: Model identifier
Returns:
FAISS index and mapping from FAISS indices to document indices
"""
# single queue in case a gpu fails
input_queue = mp.Queue()
output_queue = mp.Queue()
workers = []
for gpu_id in range(num_gpus):
worker = FastEmbedder(
gpu_id,
input_queue,
output_queue,
model_id=model_id,
batch_size=batch_size,
embedding_size=embedding_size
)
worker.start()
workers.append(worker)
index = faiss.IndexFlatIP(embedding_size)
faiss_to_doc_idx = {}
current_faiss_idx = 0
# Distribute work
for i in range(0, len(doc_list), batch_size * num_gpus):
for gpu_id in range(num_gpus):
start_idx = i + (gpu_id * batch_size)
end_idx = min(start_idx + batch_size, len(doc_list))
if start_idx >= len(doc_list):
break
batch_docs = doc_list[start_idx:end_idx]
input_queue.put({
'texts': batch_docs,
'doc_indices': list(range(start_idx, end_idx))
})
# Send stop signals and collect results
for q in workers:
input_queue.put(None)
completed = 0
while completed < num_gpus:
result = output_queue.get()
if result is None:
completed += 1
continue
index.add(result['embeddings'])
for doc_idx in result['doc_indices']:
faiss_to_doc_idx[current_faiss_idx] = doc_idx
current_faiss_idx += 1
faiss.write_index(index, 'embeddings.index')
with open('faiss_to_doc_idx.pkl', 'wb') as f:
pickle.dump(faiss_to_doc_idx, f)
return index, faiss_to_doc_idx
def create_document_list_with_relations(dataset: Dataset) -> Tuple[List[str], Dict[str, int], Dict[int, str], Dict[int, Dict[str, set]]]:
"""Create document lists and relationship mappings from dataset.
Args:
dataset: HuggingFace dataset containing query-positive-negative triplets
Returns:
Tuple containing:
- List of unique documents
- Document to index mapping
- Index to document mapping
- Query relationship mapping
"""
unique_docs = set()
query_relations = {}
for batch in dataset:
if not (batch['query'] == "" or batch['positive'] == "" or batch['negative'] == ""):
unique_docs.update([batch['query'], batch['positive'], batch['negative']])
if batch['query'] not in query_relations:
query_relations[batch['query']] = {
'positive': set(),
'negative': set()
}
query_relations[batch['query']]['positive'].add(batch['positive'])
query_relations[batch['query']]['negative'].add(batch['negative'])
doc_list = sorted(list(unique_docs))
doc_to_idx = {doc: idx for idx, doc in enumerate(doc_list)}
idx_to_doc = {idx: doc for idx, doc in enumerate(doc_list)}
query_idx_relations = {}
for query, relations in query_relations.items():
query_idx = doc_to_idx[query]
query_idx_relations[query_idx] = {
'positive': {doc_to_idx[pos] for pos in relations['positive']},
'negative': {doc_to_idx[neg] for neg in relations['negative']}
}
return doc_list, doc_to_idx, idx_to_doc, query_idx_relations
def create_hardneg_dataset(
query_idx_relations: Dict[int, Dict[str, set]],
hard_negatives: Dict[int, int],
doc_list: List[str]
) -> Dataset:
"""Create dataset with hard negatives.
Args:
query_idx_relations: Query relationship mapping
hard_negatives: Mapping of queries to hard negative indices
doc_list: List of documents
Returns:
HuggingFace dataset with query-positive-hardnegative triplets
"""
new_dataset = []
for query_idx, relations in query_idx_relations.items():
if query_idx in hard_negatives:
new_example = {
'query': doc_list[query_idx],
'positive': doc_list[list(relations['positive'])[0]],
'negative': doc_list[hard_negatives[query_idx]]
}
new_dataset.append(new_example)
return Dataset.from_dict({
'query': [ex['query'] for ex in new_dataset],
'positive': [ex['positive'] for ex in new_dataset],
'negative': [ex['negative'] for ex in new_dataset]
})
def create_parallel_hard_negatives(
index: faiss.IndexFlatIP,
faiss_to_doc_idx: Dict[int, int],
doc_list: List[str],
query_idx_relations: Dict[int, Dict[str, set]],
num_gpus: int = 8,
rerank: bool = True,
save_both: bool = True
faiss_gpu: bool = True,
save_name: str = "embeddings_huge_hardneg_dataset_rerank",
model_id: Union[str, None] = "Alibaba-NLP/gte-multilingual-reranker-base",
k_hard: int = 1,
random_n: int = 0,
) -> Dict[int, int]:
"""
Create hard negative examples using parallel GPU processing with FAISS.
Args:
index: FAISS index containing document embeddings
faiss_to_doc_idx: Mapping from FAISS indices to document indices
doc_list: List of document texts
query_idx_relations: Dictionary mapping query indices to their positive/negative relations
num_gpus: Number of GPUs to use for parallel processing
rerank: Whether to use reranking on candidates
rerank: save both rerank + non-rerank
faiss_gpu: Whether to use GPU acceleration for FAISS
save_name: Name for saving the resulting dataset
model_id: Name of HF Reranker model
k_hard: the number of hard negatives to find for each query
random_n: the number of random negatives to add to each query
Returns:
Dict[int, int]: Mapping of query indices to their hard negative indices
The function:
1. Distributes work across multiple GPUs
2. Finds hard negatives using FAISS similarity search
3. Optionally reranks candidates
4. Creates and saves a dataset with the hard negatives
"""
input_queue = mp.Queue()
output_queue = mp.Queue()
workers = []
for gpu_id in range(num_gpus):
worker = ParallelFaissSearcher(
gpu_id,
input_queue, output_queue,
index,
faiss_to_doc_idx,
doc_list,
query_idx_relations,
rerank,
model_id=model_id,
faiss_use_gpu=faiss_gpu
k_hard=k_hard
)
workers.append(worker)
query_indices = list(query_idx_relations.keys())
batch_size = len(query_indices) // (num_gpus * 4)
for i in range(0, len(query_indices), batch_size):
gpu_id = (i // batch_size) % num_gpus
batch = query_indices[i:i + batch_size]
input_queue.put(batch)
for _ in workers:
input_queue.put(None)
for worker in workers:
worker.start()
hard_negatives = {}
hard_negatives_rerank = {}
completed = 0
while completed < num_gpus:
result = output_queue.get()
if result is None:
completed += 1
continue
if rerank:
key = list(result.keys())[0]
result[key][1]
hard_negatives_rerank[key] = result[key][1]
hard_negatives[key] = result[key][0]
else:
hard_negatives.update(result)
if rerank:
if save_both:
hardneg_dataset = create_hardneg_dataset(
query_idx_relations, hard_negatives, doc_list, random_n
)
hardneg_dataset.save_to_disk(save_name)
hardneg_dataset = create_hardneg_dataset(
query_idx_relations, hard_negatives_rerank, doc_list, random_n
)
hardneg_dataset.save_to_disk(save_name + '_rerank')
else:
hardneg_dataset = create_hardneg_dataset(
query_idx_relations, hard_negatives, doc_list, random_n
)
hardneg_dataset.save_to_disk(save_name)
return hard_negatives
def create_aligned_datasets(datasets_dict: DatasetDict) -> DatasetDict:
"""Create aligned datasets using ordered hash-based matching."""
# Get the smallest dataset as reference
smallest_dataset = min(datasets_dict.values(), key=len)
# Create ordered master list of hash pairs from smallest dataset
master_hashes = [(hashlib.sha256(q.encode()).hexdigest(),
hashlib.sha256(p.encode()).hexdigest())
for q, p in zip(smallest_dataset['query'],
smallest_dataset['positive'])]
# Find matching indices in each dataset that preserve order
aligned_datasets = {}
for name, dataset in datasets_dict.items():
current_hashes = [(hashlib.sha256(q.encode()).hexdigest(),
hashlib.sha256(p.encode()).hexdigest())
for q, p in zip(dataset['query'], dataset['positive'])]
# Keep track of which master hashes we've found
found_indices = []
for master_hash in master_hashes:
if master_hash in current_hashes:
idx = current_hashes.index(master_hash)
found_indices.append(idx)
aligned_datasets[name] = dataset.select(found_indices)
def verify_alignment():
print("\nAlignment Statistics:")
sizes = [len(dataset) for dataset in aligned_datasets.values()]
print(f"All datasets aligned to size: {sizes[0]}")
assert all(size == sizes[0] for size in sizes), "Dataset sizes don't match"
base_hashes = [(hashlib.sha256(q.encode()).hexdigest(),
hashlib.sha256(p.encode()).hexdigest())
for q, p in zip(list(aligned_datasets.values())[0]['query'],
list(aligned_datasets.values())[0]['positive'])]
for dataset in aligned_datasets.values():
current_hashes = [(hashlib.sha256(q.encode()).hexdigest(),
hashlib.sha256(p.encode()).hexdigest())
for q, p in zip(dataset['query'], dataset['positive'])]
assert base_hashes == current_hashes, "Hash sequences don't match exactly"
verify_alignment()
return aligned_datasets
def save_aligned_data(aligned_splits: dict, base_path: str) -> None:
"""Save each dataset as a DatasetDict to disk."""
for dataset_name, dataset in aligned_splits.items():
save_path = f"{base_path}/{dataset_name}"
dataset.save_to_disk(save_path)
Essentially, we create a queue/job process that attempts to overload the model/faiss bottlenecks. We allocate a number of processes to our gpus where n_gpus == n_processes. You have to know size of model/datasizing so you’re not having your gpu processes compete for non-existant memory (eg 20 processes while only having 8 GPUs), so back-of-the-napkin math means just keep it simple and set n_processes equal to n_gpus.
We multiprocess on each gpu in our stack (8x) and first find hard-negatives via BM25, then a model alone, then a model + re-ranker.
Here is an image of what it looks like (or at least what I hope to see), that the reranker will provide the best result for the given query - caveat: to the astute reader, we’re using an untuned reranker (eg not tuned for law data) - so I don’t expect it to be the best, I just expect it to do better than average. But in reality, I have no real idea what to expect, I sort of just want to ‘see with my own eyes what happens’ when we play around with various hard-negative mining strategies.
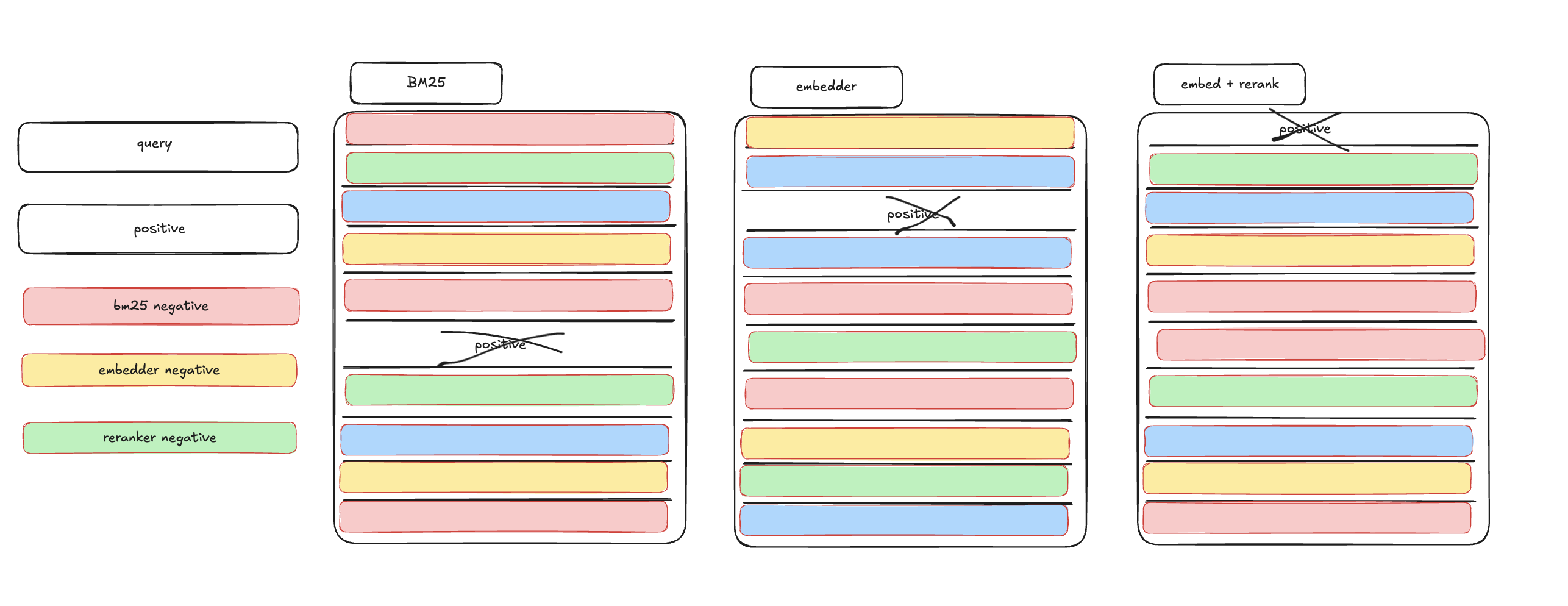
ok, less pontificating, more code:
Define our dataset/model configs
dataset_config = {
"name": "joelniklaus/legal_case_document_summarization",
"splits": ["train", "test"],
"pairs": [("judgement", "summary")]
}
num_gpus=8
batch_size=1
embedding_size=1536
model_id = "Alibaba-NLP/gte-Qwen2-1.5B-instruct"
Create our datasets
# create our base bm25 dataset
bm25_dataset = create_bm25_data(dataset_config, save_name="legal_case_document_summarization_bm25")
# create our faiss index using our main
doc_list, doc_to_idx, idx_to_doc, query_idx_relations = create_document_list_with_relations(bm25_dataset)
index, faiss_to_doc_idx = create_fast_index(
doc_list, num_gpus=num_gpus, batch_size=batch_size,
embedding_size=embedding_size, model_id=model_id
)
_ = create_parallel_hard_negatives("embeddings.index", faiss_to_doc_idx, doc_list, query_idx_relations, num_gpus=num_gpus, rerank=True, save_both=True, faiss_gpu=True, save_name="legal_case_document_summarization", k_hard=1, random_n=0)
(our datasets will be created at this point.)
super simple training script
import argparse
from datasets import load_from_disk
from sentence_transformers import (
SentenceTransformer,
SentenceTransformerTrainer,
SentenceTransformerTrainingArguments,
)
from sentence_transformers.evaluation import TripletEvaluator
from sentence_transformers.losses import CachedMultipleNegativesRankingLoss
from sentence_transformers.training_args import BatchSamplers
import torch
torch.set_float32_matmul_precision('high')
def train(dataset_path, dataset_name):
torch.set_float32_matmul_precision('high')
# parse the lr & model name
lr = 8e-5
model_name = "answerdotai/ModernBERT-base"
model_shortname = model_name.split("/")[-1]
# 1. Load a model to finetune
model = SentenceTransformer(model_name)
# 2. Load a dataset to finetune on
train_dataset = load_from_disk(dataset_path)
# 3. Define a loss function
loss = CachedMultipleNegativesRankingLoss(model, mini_batch_size=16) # Increase mini_batch_size if you have enough VRAM
run_name = f"{model_shortname}-{dataset_name}"
# 4. (Optional) Specify training arguments
args = SentenceTransformerTrainingArguments(
# Required parameter:
output_dir=f"output/{model_shortname}/{run_name}",
# Optional training parameters:
num_train_epochs=1,
per_device_train_batch_size=2,
per_device_eval_batch_size=2,
warmup_ratio=0.05,
fp16=False, # Set to False if GPU can't handle FP16
bf16=True, # Set to True if GPU supports BF16
batch_sampler=BatchSamplers.NO_DUPLICATES, # (Cached)MultipleNegativesRankingLoss benefits from no duplicates
learning_rate=lr,
# Optional tracking/debugging parameters:
save_strategy="steps",
save_steps=200,
save_total_limit=2,
logging_steps=200,
run_name=run_name, # Used in `wandb`, `tensorboard`, `neptune`, etc. if installed
)
# 6. Create a trainer & train
trainer = SentenceTransformerTrainer(
model=model,
args=args,
train_dataset=train_dataset,
loss=loss,
)
trainer.train()
# 8. Save the model
model.save_pretrained(f"output/{run_name}/final")
if __name__ == '__main__':
parser = argparse.ArgumentParser()
parser.add_argument("--dataset_path", type=str)
parser.add_argument("--dataset_name", type=str)
args = parser.parse_args()
train(args.dataset_path, args.dataset_name)
the unresonable effectiveness of bm25?
In my opinion, there’s no real edge here to anyone - we can discount AILA/LegalQuAD for now - and notice that in the other three tasks, each method (bm25, rerank, no-rerank) gets one victory a piece.
As for the foreign legal data, bm25 does a better on two indian legal tasks, but on the german task, bm25 is the worst.
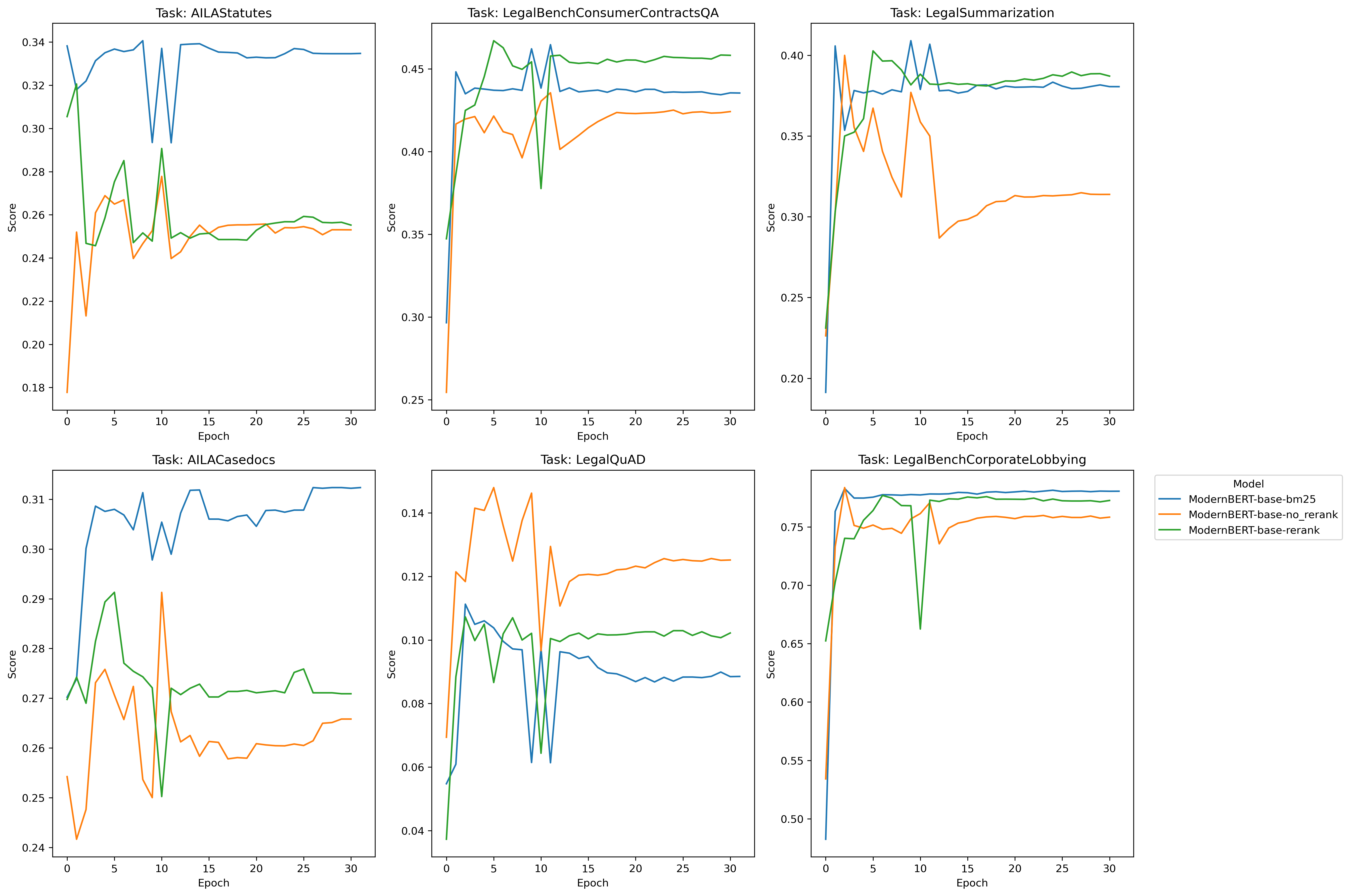
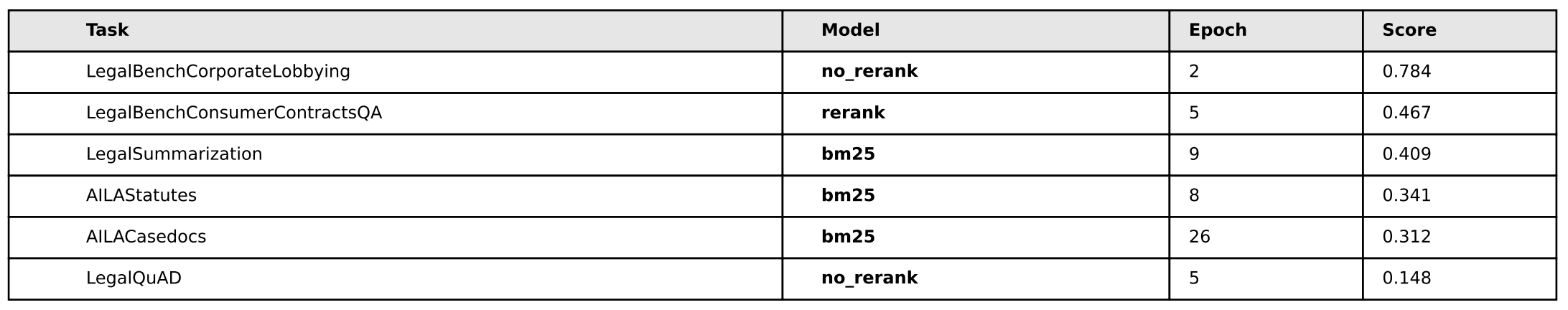
I do notice one interesting thing on all tasks, however, and that’s rerank and no-rerank seem to be inversely correlated on certain performance spikes: when rerank goes up, no-rerank goes down and vise-versa - so let’s see what happens when we create combinations of these pairs and train with that data and see what happens:
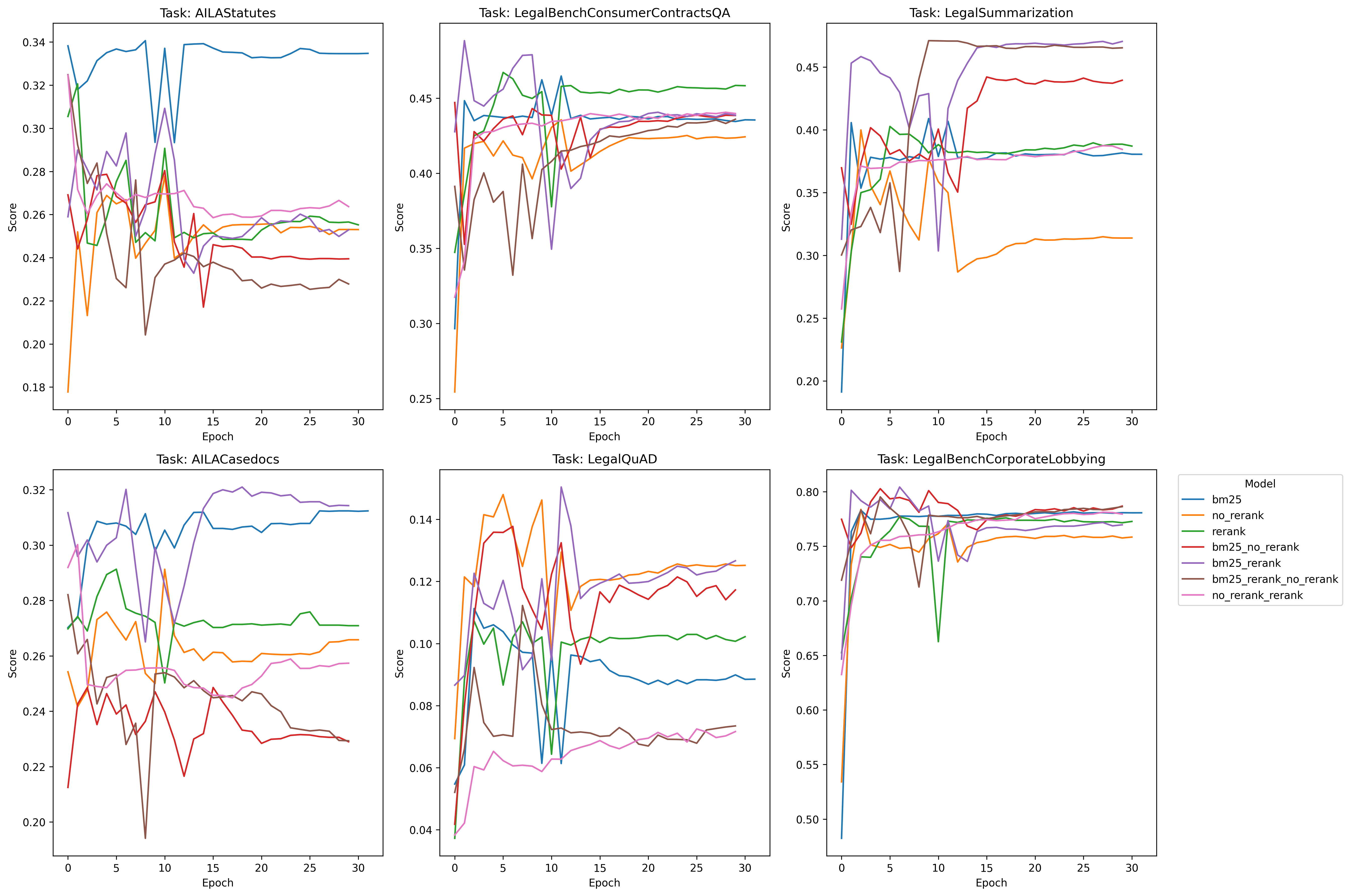
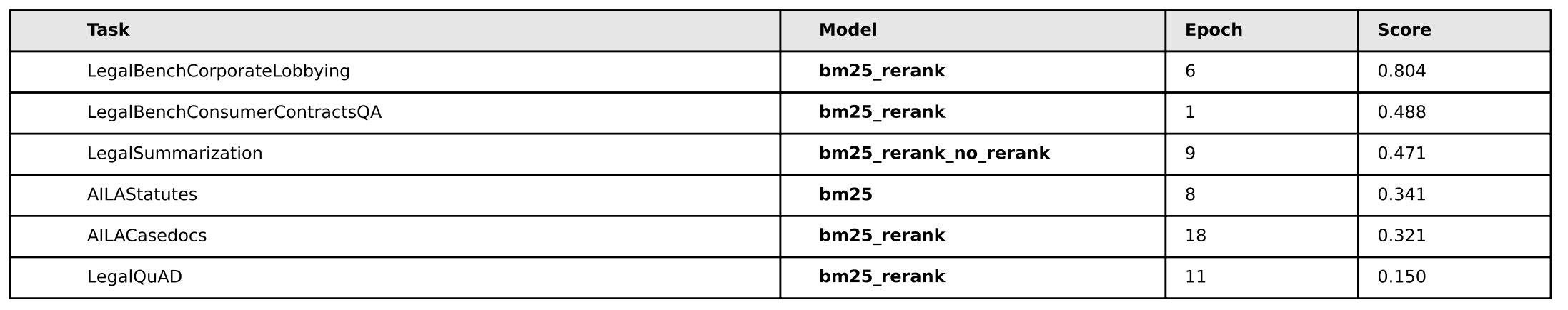
Cool! Much better performance. It seems that some combination of the simplicity of bm25 + the rerank method yield superior performance in all areas except Legal Summarization - which is interesting considering our training dataset is exclusively legal summarization data, so contextually I suppose that sort of makes sense. We don’t know however if this was due exclusively to having more negative examples per query, so we’ll have to test with random data (1 vs. 2. vs. 3, etc) and see if it’s just the presence of an extra example that makes the difference – we’ll test on that later.
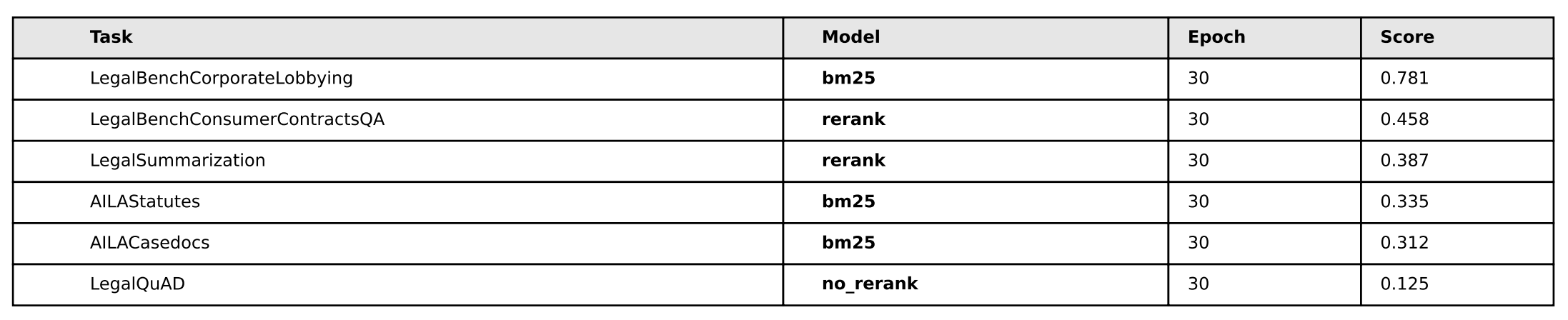
The other idea I had was to let these models overtrain for many epochs ‘just to see what happens’. So you’ll notice that in all the models, they’re trained for 30 epochs when that’s probably far, far too much. The reason for this is I sort of had a hunch that because bm25 is “semantically simpler”, it should be the ‘most resistant to overfitting’ as compared to the other two models. So I wanted to see if the model would overfit to the training data and then what would happen if we let it overtrain for a long time. It turned out that’s not necessarily the case, rerank actually performs better than bm25 on the subtasks we’re most curious about.
Also, and some what serenditiously, based on the observations a poster made on a random twitter thread, I was curious what would happen if we average/ensemble the weights of the three ((bm25 + rerank + no-rerank) / 3) at each checkpoint and measure the perf:
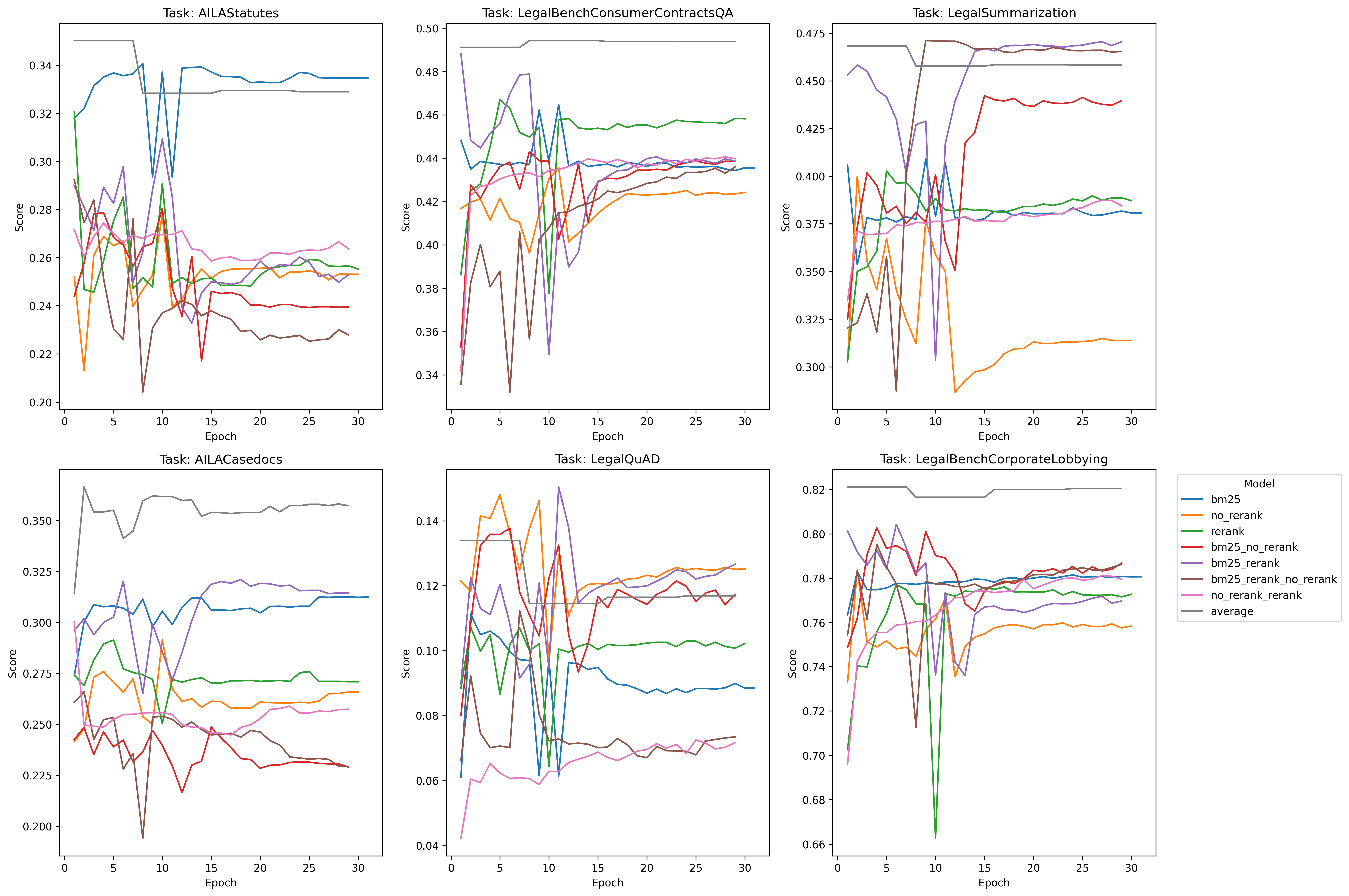
Much better on the foreign legal tasks - and the take away is probably that if you’re compute constrained and want the best performing model, this is probably your best bet all things considered: simply take the average of various data-mixes. It kind of goes to show that implicit regularization is so unbelievably powerful that it should probably be one of your first go to perf increase tools.
Intuitively, the way I’ve been thinking about this is that bm25 picked hard-negatives alone is probably some form of regularization in some simplistic capacity.
A few interesting rabit hole ideas
Looking at the training curves, an idea I had was to see if we could somehow use the stability of bm25 and use varying degrees of noise/hard-negatives to get more lift in performance - as well as test varying amounts of hard-negatives per query:
what impact does bm25 have when it’s supplemented by specifically chosen vs random hardnegs;
what if we progressively add more hard-negatives as the training steps move forward (epoch 1: 1 hardneg, epoch 2: 2 hardnegs, … epoch 2000: 2000 hardnegs),
we make it, theoretically more difficult/different looks as training continues: either by always showing the model a new batch of hard-negatives, or by increasing the number of hard-negatives per batch?
At this point I’m sort of just trying things based on the loss-curves/eval scores - like I notice that bm25 is sort of resistant to overtraining or at least has good stable training grad-norms, so I sort of just picked a random idea to start and let the research muse take me where it meant to take me after that.
CODE
I started writing one comprehensive/thoughtful code base, but when the research god whispers, you hack something together and just run the script.
Below are all the pieces of code for all the random experiments I ran in no particular order::
Random
At the end of each epoch, we take each sample (query, positive) in our dateset and we randomly chose a hard-negative to pair with the query/positive pair. (this method did horribly)
#pseudo code:
def on_epoch_end(self, **kwargs):
for qid, relations in self.query_idx_relations:
cache_entries = self.query_caches[qid]
current_len = len(cache_entries)
if current_len < self.k_cache:
needed = self.k_cache - current_len
# Example: random pick from global_pool
import random
candidates = set(self.idx_to_doc) - set(relations['positive'])
new_candidates = random.sample(candidates, min(len(candidates), needed))
# Compute hardness for each new candidate
q_text = self.doc_list[qid]
pos_text = self.doc_list[list(relations['positive'])[0]]
new_entries = []
for cand_idx in new_candidates:
cand_text = self.doc_list[cand_idx]
new_entries.append(cand_text)
self.query_caches[qid].extend(new_entries)
....
Model-Based Mining:
At the end of each epoch (or after some time, we can set the start time with ‘epoch_start’), for each piece of text w/in our dataset ({query1, query2, ..,positive1, positive2, queryN, positiveN}), we take the current model and we embed the entire dataset. For each original query/positive pair, we find the K most similar texts for each query that isn’t it’s corresponding positive. We add some parameters to select the number K and if we want to add random hard-negatives as noise.
class HardNegMiningCallback(TrainerCallback):
"""
A callback that, at the end of each epoch:
- If we're on the main process, sets CUDA_VISIBLE_DEVICES to '6,7'
- Calls create_hardnegs(...)
- Waits for completion
- Reloads the dataset with newly generated negatives
"""
def __init__(
self,
trainer, # <--- pass in the trainer itself
create_hardnegs_fn,
original_dataset_path,
adaptive_dataset_path,
model_path=None,
gpus_for_hardneg="6,7",
reload_after=True,
num_gpus_for_hardneg=2,
batch_size=256,
embedding_size=1536,
epoch_start=1,
k_hard=1,
random_n=0,
):
"""
Args:
trainer: The HF Trainer (or a subclass) that we want to manipulate.
create_hardnegs_fn: A function with signature
create_hardnegs(dataset_path, save_path, model_path, num_gpus, batch_size, embedding_size)
original_dataset_path: path to the dataset we started training on
adaptive_dataset_path: where we save the newly created dataset with fresh negatives
model_path: path to the current model or checkpoint (trainer.args.output_dir by default)
gpus_for_hardneg: which GPUs (by index) to use for negative generation (e.g. "6,7")
reload_after: if True, reload the newly generated dataset into trainer.train_dataset
num_gpus_for_hardneg: how many GPUs we actually want to use inside create_hardnegs
batch_size, embedding_size: additional parameters to pass to create_hardnegs
epoch_start: only start doing negative generation after this epoch number
k_hard: number of hard negatives to generate per query
"""
super().__init__()
self.trainer = trainer # store a reference to the trainer
self.create_hardnegs_fn = create_hardnegs_fn
self.original_dataset_path = original_dataset_path
self.adaptive_dataset_path = adaptive_dataset_path
self.model_path = model_path
self.gpus_for_hardneg = gpus_for_hardneg
self.reload_after = reload_after
self.num_gpus_for_hardneg = num_gpus_for_hardneg
self.batch_size = batch_size
self.embedding_size = embedding_size
self.epoch_start = epoch_start
self.k_hard = k_hard
self.random_n = random_n
self.epoch_counter = 0
def on_epoch_end(
self,
args,
state: TrainerState,
control: TrainerControl,
**kwargs
):
self.epoch_counter += 1
if self.epoch_counter < self.epoch_start:
return # skip until epoch_start is reached
# Determine if we're rank=0 in distributed training
# Hugging Face sets this in TrainerState or we can check:
# if self.trainer.is_world_process_zero():
# or
# if state.is_world_process_zero
# (They generally do the same in modern versions.)
is_main_process = getattr(self.trainer, "is_world_process_zero", None)
if callable(is_main_process):
is_main_process = self.trainer.is_world_process_zero()
elif hasattr(state, "is_world_process_zero"):
is_main_process = state.is_world_process_zero
else:
# Fallback: assume single process or assume main
is_main_process = True
if is_main_process:
print(f"\n[HardNegMiningCallback] Epoch {self.epoch_counter} ended. Creating new hard negatives on GPUs {self.gpus_for_hardneg}.")
# 1) Save the original CUDA_VISIBLE_DEVICES so we can restore it later
original_cuda_env = os.environ.get("CUDA_VISIBLE_DEVICES", "")
print(f"[HardNegMiningCallback] Original CUDA_VISIBLE_DEVICES: {original_cuda_env}")
try:
# 2) Set env so that only GPUs 6,7 are visible for negative generation
os.environ["CUDA_VISIBLE_DEVICES"] = self.gpus_for_hardneg
print(f"[HardNegMiningCallback] Setting CUDA_VISIBLE_DEVICES to {os.environ['CUDA_VISIBLE_DEVICES']}")
# 3) Determine the current model checkpoint path
model_ckpt_path = self.model_path or self.trainer.args.output_dir
# we get the final checkpoint h
checkpoints = [d for d in os.listdir(model_ckpt_path) if d.startswith('checkpoint-')]
# Sort checkpoints by epoch number
checkpoints.sort(key=lambda x: int(x.split('-')[1]))
# get last checkpoint
checkpoint_path = os.path.join(model_ckpt_path, checkpoints[-1])
# 4) Call the user-provided function
self.create_hardnegs_fn(
dataset_path=self.original_dataset_path,
save_path=self.adaptive_dataset_path,
model_path=checkpoint_path,
num_gpus=self.num_gpus_for_hardneg,
batch_size=self.batch_size,
embedding_size=self.embedding_size,
k_hard=self.k_hard,
random_n=self.random_n,
)
finally:
# 5) Restore original CUDA_VISIBLE_DEVICES for training
os.environ["CUDA_VISIBLE_DEVICES"] = original_cuda_env
print(f"[HardNegMiningCallback] Restored CUDA_VISIBLE_DEVICES to {os.environ['CUDA_VISIBLE_DEVICES']}")
# 6) Optional: Wait for all ranks if in distributed mode
if dist.is_initialized():
dist.barrier()
# 7) Reload the updated dataset for the next epoch
if self.reload_after:
print(f"[HardNegMiningCallback] Reloading dataset from {self.adaptive_dataset_path}")
updated_dataset = load_from_disk(self.adaptive_dataset_path)
self.trainer.train_dataset = updated_dataset
# Some versions store internally in `_train_dataset`
self.trainer._train_dataset = updated_dataset
self.trainer.train_dataloader = self.trainer.get_train_dataloader()
# If you want to keep updating original_dataset_path to the new path each time
self.original_dataset_path = self.adaptive_dataset_path
def build_new_training_lr_schedule(self, new_dataset):
current_lr = self.trainer.optimizer.param_groups[0]['lr']
# Calculate new total steps based on new dataset size
num_training_steps = (
len(new_dataset)
* self.trainer.args.num_train_epochs
// (self.trainer.args.per_device_train_batch_size * self.trainer.args.gradient_accumulation_steps)
)
# Create new scheduler starting from current lr
# update the scheduler to accompany the new size
new_scheduler = get_cosine_schedule_with_warmup(
self.trainer.optimizer,
num_warmup_steps=0, # No warmup since we're continuing
num_training_steps=num_training_steps,
initial_lr=current_lr
)
self.trainer.lr_scheduler = new_scheduler
Margin Based Random Mining:
So in this version, we do the same model-based mining as before, but we add a margin parameter to compute whether the model has ‘learned’ the example or not - and instead of removing the triplet prematurely, we keep continually showing the model the same triplet if the margin is over a certain threshold, if not, we add a new randomly selected hard-negative. The thgouht here is want to keep only truly hard negatives in the cache and vacate those that the model learns.
class AdaptiveHardNegMiningCallback(TrainerCallback):
"""
Maintains a per-query cache of negatives. On each epoch end (or step end), it:
- samples from the cache for training
- updates each query's cache by removing "easy" negatives
- periodically refills from a global pool or partial re-embedding
"""
def __init__(
self,
trainer, # reference to the Trainer (or your model)
train_dataset, # list of query texts or IDs
model, # the retrieval model or sentence-transformer
k_cache=5, # how many negatives we keep in each cache
remove_threshold=-0.05, # if margin < -0.05 => negative is "easy"
refill_every=2, # every 2 epochs, we try to refill or re-embed
create_hardnegs_fn=None,
original_dataset_path=None,
adaptive_dataset_path=None,
model_path=None,
gpus_for_hardneg="6,7",
reload_after=True,
num_gpus_for_hardneg=2,
batch_size=256,
embedding_size=1536,
epoch_start=1,
k_hard=1,
random_n=0,
):
super().__init__()
self.trainer = trainer
self.model = model
#self.global_pool = global_pool or []
self.k_cache = k_cache
self.remove_threshold = remove_threshold
self.refill_every = refill_every
self.create_hardnegs_fn = create_hardnegs_fn
self.original_dataset_path = original_dataset_path
self.adaptive_dataset_path = adaptive_dataset_path
self.model_path = model_path
self.gpus_for_hardneg = gpus_for_hardneg
self.reload_after = reload_after
self.num_gpus_for_hardneg = num_gpus_for_hardneg
self.batch_size = batch_size
self.embedding_size = embedding_size
self.epoch_start = epoch_start
self.k_hard = k_hard
self.random_n = random_n
# Build initial caches
# Each cache is a list of (neg_text, hardness) or just neg_text
self.doc_list, self.doc_to_idx, self.idx_to_doc, self.query_idx_relations = create_document_list_with_relations(train_dataset)
# alld
self.query_caches = {}
for qid in self.query_idx_relations:
# pick up to k_cache from initial_negatives
negs = self.query_idx_relations['negatives'][:k_cache]
self.query_caches[qid] = [(n, 999.0) for n in negs] # hardness placeholder, # (int, int)
self.epoch_counter = 0
def on_epoch_begin(self, args, state: TrainerState, control: TrainerControl, **kwargs):
"""
Optionally, we can sample a brand-new training dataset from caches here.
Or do it on_step_end if you prefer a finer granularity.
"""
# We can rebuild or shuffle the "train_dataset" from these caches
self._build_train_dataset_from_caches()
# No direct return needed, the trainer will continue
def on_epoch_end(
self,
args,
state: TrainerState,
control: TrainerControl,
**kwargs
):
self.epoch_counter += 1
# 1) Update caches for each query -> remove easy negatives
self._update_caches_remove_easy()
# 2) Periodically refill caches from global pool or partial re-embedding
if (self.epoch_counter % self.refill_every) == 0:
self._refill_caches()
# 3) Optionally: re-build training dataset from updated caches
# so next epoch sees the new negatives
self._build_train_dataset_from_caches()
################################################################
# Internals
################################################################
def _build_train_dataset_from_caches(self):
"""
Rebuild a 'training dataset' from the caches if we want to feed it
into Trainer. We can do something like:
train_dataset = List of InputExample(query, pos, neg)
Then: self.trainer.train_dataset = <some huggingface Dataset>
"""
new_samples = []
for qid, relations in self.query_idx_relations:
cache_entries = self.query_caches[qid]
for (neg_idx, hardness) in cache_entries:
new_example = {
'query': self.doc_list[qid],
'positive': self.doc_list[list(relations['positive'])[0]],
'negative': self.doc_list[neg_idx]
}
new_samples.append(new_example)
# Now wrap that in a HF Dataset, or a custom dataset for your trainer
# For quick pseudo-code:
from datasets import Dataset
train_dataset = Dataset.from_list(new_samples)
self.trainer.train_dataset = train_dataset
self.trainer._train_dataset = train_dataset # if needed
# You might want to re-init dataloader:
self.trainer.train_dataloader = self.trainer.get_train_dataloader()
def _update_caches_remove_easy(self):
"""
For each (query, neg), check if it's now 'easy' for the model:
- if margin < remove_threshold => remove from the cache
- otherwise, keep it (it's still hard)
"""
for qid, relations in self.query_idx_relations:
new_cache = []
q_text = self.doc_list[qid]
pos_text = self.doc_list[list(relations['positive'])[0]]
for (neg_id, old_hardness) in self.query_caches[qid]:
neg_text = self.doc_list[neg_id]
margin = compute_hardness(self.model, q_text, pos_text, neg_text)
if margin < self.remove_threshold:
# It's become easy => we remove it
pass
else:
# It's still hard, keep it
new_cache.append((neg_text, margin))
self.query_caches[qid] = new_cache
def _refill_caches(self):
"""
If the caches have fewer than k_cache negatives, we pick new ones from
the global pool or re-embed & find new top-K. For simplicity, let's do random
from global_pool here.
"""
# can do re-embed here
for qid, relations in self.query_idx_relations:
cache_entries = self.query_caches[qid]
current_len = len(cache_entries)
if current_len < self.k_cache:
needed = self.k_cache - current_len
# Example: random pick from global_pool
import random
candidates = set(self.idx_to_doc) - set(relations['positive'])
new_candidates = random.sample(candidates, min(len(candidates), needed))
# Compute hardness for each new candidate
q_text = self.doc_list[qid]
pos_text = self.doc_list[list(relations['positive'])[0]]
new_entries = []
for cand_idx in new_candidates:
cand_text = self.doc_list[cand_idx]
margin = compute_hardness(self.model, q_text, pos_text, cand_text)
new_entries.append((cand_text, margin))
self.query_caches[qid].extend(new_entries)
def build_new_training_lr_schedule(self, new_dataset):
current_lr = self.trainer.optimizer.param_groups[0]['lr']
# Calculate new total steps based on new dataset size
num_training_steps = (
len(new_dataset)
* self.trainer.args.num_train_epochs
// (self.trainer.args.per_device_train_batch_size * self.trainer.args.gradient_accumulation_steps)
)
# Create new scheduler starting from current lr
# update the scheduler to accompany the new size
new_scheduler = get_cosine_schedule_with_warmup(
self.trainer.optimizer,
num_warmup_steps=0, # No warmup since we're continuing
num_training_steps=num_training_steps,
initial_lr=current_lr
)
self.trainer.lr_scheduler = new_scheduler
Margin Based Bandit Mining:
In this version, we combine both the original model-based mining and the margin-based mining and instead of subjectively picking a margin threshold to empty the hard-negative cache for a given sample, we use an Epsilon Greedy Bandit to pick the margin value. This one is cool, so I’m patting myself on the back for this.
Epsilon Greedy knowledge check: we provide a list of arms where at each index is the margin threshold that we’ve assigned to that arm, we also provide an epsilon value - which is the probability for exploring instead of taking the highest reward, and an initial reward of 0 for each arm. On the epoch’s start, we use the select arm function which picks the given arm either randomly based on epsilon or best performing arm (the one with the max qvalues).
On the epoch end, we update the q-value for the chosen arm based on the reward from our eval set.
class EpsilonGreedyBandit:
"""
Each arm is a candidate threshold.
We track (mean_reward, count) for each arm.
We pick arms via epsilon-greedy.
"""
def __init__(self, arms, epsilon=0.1, initial_value=0.0):
"""
:param arms: list of thresholds (floats)
:param epsilon: probability of exploring
:param initial_value: initial assumed reward for each arm
"""
self.arms = arms
self.epsilon = epsilon
self.q_values = [initial_value for _ in arms] # estimated mean reward
self.counts = [0 for _ in arms] # how many times each arm was used
def select_arm(self):
"""
Epsilon-greedy selection.
"""
if random.random() < self.epsilon:
# Explore
return random.randint(0, len(self.arms)-1)
else:
# Exploit (pick best so far)
return int(np.argmax(self.q_values))
def update(self, arm_index, reward):
"""
Update estimates for the chosen arm.
"""
self.counts[arm_index] += 1
c = self.counts[arm_index]
old_q = self.q_values[arm_index]
# incremental mean update
new_q = old_q + (reward - old_q) / c
self.q_values[arm_index] = new_q
def get_arm_value(self, arm_index):
return self.arms[arm_index]
def __repr__(self):
return f"EpsilonGreedyBandit(arms={self.arms}, q_values={self.q_values}, counts={self.counts})"
class AdaptiveBanditHardNegMiningCallback(TrainerCallback):
"""
Maintains a per-query cache of negatives.
Uses a bandit to pick 'remove_threshold' at the start of each epoch.
"""
def __init__(
self,
trainer, # reference to the Trainer
train_dataset,
model,
k_cache=5,
bandit_arms=None, # list of candidate thresholds
epsilon=0.1,
refill_every=2,
eval_metric_key="eval_loss" # or "eval_accuracy", "eval_mrr", etc.
):
super().__init__()
self.trainer = trainer
self.model = model
self.k_cache = k_cache
self.refill_every = refill_every
self.eval_metric_key = eval_metric_key
# 1) Build bandit
if bandit_arms is None:
# Provide default arms if not specified
bandit_arms = [-0.1, -0.05, 0.0, 0.05, 0.1]
self.bandit = EpsilonGreedyBandit(arms=bandit_arms, epsilon=epsilon)
# 2) Build initial caches, etc. (similar to your original code)
self.doc_list, self.doc_to_idx, self.idx_to_doc, self.query_idx_relations = create_document_list_with_relations(train_dataset)
self.query_caches = {}
for qid in self.query_idx_relations:
negs = list(self.query_idx_relations[qid]['negative']) # example
# pick up to k_cache
chosen = negs[:k_cache]
self.query_caches[qid] = [(nid, 999.0) for nid in chosen]
self.epoch_counter = 0
self.current_arm_index = None # which threshold arm we used this epoch
self.remove_threshold = None # the actual threshold for this epoch
def on_epoch_begin(self, args, state: TrainerState, control: TrainerControl, **kwargs):
"""
1) Pick an arm from bandit => set remove_threshold
2) Rebuild the training dataset
"""
self.epoch_counter += 1
# (A) Bandit chooses which arm to use
self.current_arm_index = self.bandit.select_arm()
self.remove_threshold = self.bandit.get_arm_value(self.current_arm_index)
print(f"[BanditCallback] Epoch={self.epoch_counter}, chosen threshold={self.remove_threshold:.3f}")
# (B) Re-build / shuffle training data
self._build_train_dataset_from_caches()
def on_epoch_end(self, args, state: TrainerState, control: TrainerControl, **kwargs):
"""
1) Update caches => remove easy negatives
2) Optionally refill
3) Evaluate => measure reward => update bandit
"""
# 1) Remove easy negatives
self._update_caches_remove_easy_batch()
# 2) Refill if needed
if (self.epoch_counter % self.refill_every) == 0:
self._refill_caches_batch()
# 3) Evaluate => compute reward
# We can do a trainer.evaluate(...) or rely on existing results in state.log_history
# For example, let's see if we have an eval result in state.log_history
# We'll find the last logged eval_metric_key
reward = 0.0
for entry in reversed(state.log_history):
if self.eval_metric_key in entry:
reward = entry[self.eval_metric_key]
break
print(f"[BanditCallback] Epoch={self.epoch_counter}, performance metric={reward:.4f}")
# 4) Update bandit with reward
self.bandit.update(self.current_arm_index, reward)
# Optionally re-build the dataset after removing negatives
self._build_train_dataset_from_caches()
############################################
# The same helper methods as before
############################################
def _build_train_dataset_from_caches(self):
# Rebuild a 'training dataset' from caches
new_samples = []
for qid, rels in self.query_idx_relations.items():
cache_entries = self.query_caches[qid]
q_text = self.doc_list[qid]
# pick 1 positive from rels
pos_text_id = list(rels['positive'])[0]
pos_text = self.doc_list[pos_text_id]
for (neg_id, hardness) in cache_entries:
neg_text = self.doc_list[neg_id]
new_samples.append({
"query": q_text,
"positive": pos_text,
"negative": neg_text
})
from datasets import Dataset
train_dataset = Dataset.from_list(new_samples)
self.trainer.train_dataset = train_dataset
self.trainer._train_dataset = train_dataset
self.trainer.train_dataloader = self.trainer.get_train_dataloader()
def _update_caches_remove_easy(self):
for qid, rels in self.query_idx_relations.items():
q_text = self.doc_list[qid]
pos_text_id = list(rels['positive'])[0]
pos_text = self.doc_list[pos_text_id]
new_cache = []
for (neg_id, old_hardness) in self.query_caches[qid]:
neg_text = self.doc_list[neg_id]
margin = compute_hardness(self.model, q_text, pos_text, neg_text)
# use self.remove_threshold from the bandit
if margin < self.remove_threshold:
# remove
pass
else:
new_cache.append((neg_id, margin))
self.query_caches[qid] = new_cache
def _refill_caches(self):
# just a skeleton: pick random negatives from the corpus to fill up to k_cache
import random
for qid, rels in self.query_idx_relations.items():
current_len = len(self.query_caches[qid])
if current_len < self.k_cache:
needed = self.k_cache - current_len
q_text = self.doc_list[qid]
pos_text = self.doc_list[list(rels['positive'])[0]]
# randomly sample from all doc_ids except positives or qid
all_candidates = list(set(self.doc_to_idx.values()) - {qid} - set(rels['positive']))
new_cands = random.sample(all_candidates, min(len(all_candidates), needed))
new_entries = []
for cidx in new_cands:
cand_text = self.doc_list[cidx]
m = compute_hardness(self.model, q_text, pos_text, cand_text)
new_entries.append((cidx, m))
self.query_caches[qid].extend(new_entries)
def _refill_caches_batch(self):
"""
Similar to _refill_caches, but we do all margin computations in batch mode
to reduce overhead.
"""
import random
batch_q = []
batch_p = []
batch_n = []
index_map = [] # (qid, candidate_neg_id, index_in_batch)
# 1) Gather all needed candidates from each query
for qid, rels in self.query_idx_relations.items():
current_len = len(self.query_caches[qid])
needed = self.k_cache - current_len
if needed <= 0:
continue
q_text = self.doc_list[qid]
pos_text = self.doc_list[list(rels['positive'])[0]]
# Sample from all candidates except qid or positives
all_candidates = (
set(self.doc_to_idx.values())
- {qid}
- set(rels['positive'])
)
if len(all_candidates) == 0:
continue
needed_cands = random.sample(all_candidates, min(len(all_candidates), needed))
# 2) Collect them in a batch list
for cand_idx in needed_cands:
cand_text = self.doc_list[cand_idx]
batch_q.append(q_text)
batch_p.append(pos_text)
batch_n.append(cand_text)
index_map.append((qid, cand_idx))
if not batch_q:
# nothing to refill
return
# 3) Batch compute margins in chunks
chunk_size = 256
margin_list = []
for start in range(0, len(batch_q), chunk_size):
end = start + chunk_size
q_chunk = batch_q[start:end]
p_chunk = batch_p[start:end]
n_chunk = batch_n[start:end]
margins = compute_batch_hardness(self.model, q_chunk, p_chunk, n_chunk)
margin_list.extend(margins)
# 4) Assign margins back to the caches
# index_map[i] -> (qid, cand_idx)
# margin_list[i] -> margin
for i, (qid, cand_idx) in enumerate(index_map):
margin_val = margin_list[i]
# just append to the cache
self.query_caches[qid].append((cand_idx, margin_val))
def _update_caches_remove_easy_batch(self):
# 1) Gather all (q_text, p_text, n_text) in a big list
batch_q = []
batch_p = []
batch_n = []
index_map = [] # keep track of (qid, neg_id, index_in_batch)
for qid, rels in self.query_idx_relations.items():
q_text = self.doc_list[qid]
pos_text_id = list(rels['positive'])[0]
p_text = self.doc_list[pos_text_id]
new_cache_entries = []
for idx, (neg_id, old_hardness) in enumerate(self.query_caches[qid]):
neg_text = self.doc_list[neg_id]
batch_q.append(q_text)
batch_p.append(p_text)
batch_n.append(neg_text)
index_map.append((qid, idx)) # so we know which cache entry this corresponds to
# 2) Possibly chunk it if it's very large
margin_list = []
chunk_size = 256
for start in range(0, len(batch_q), chunk_size):
end = start + chunk_size
q_chunk = batch_q[start:end]
p_chunk = batch_p[start:end]
n_chunk = batch_n[start:end]
margins = compute_batch_hardness(self.model, q_chunk, p_chunk, n_chunk)
margin_list.extend(margins)
# margin_list is now same length as index_map
# 3) Go back through index_map and remove easy
for (i, (qid, cache_idx)) in enumerate(index_map):
margin = margin_list[i]
if margin < self.remove_threshold:
# mark for removal
self.query_caches[qid][cache_idx] = None
else:
# store updated hardness
self.query_caches[qid][cache_idx] = (self.query_caches[qid][cache_idx][0], margin)
# 4) filter out removed
for qid in self.query_idx_relations:
self.query_caches[qid] = [x for x in self.query_caches[qid] if x is not None]
def build_new_training_lr_schedule(self, new_dataset):
current_lr = self.trainer.optimizer.param_groups[0]['lr']
# Calculate new total steps based on new dataset size
num_training_steps = (
len(new_dataset)
* self.trainer.args.num_train_epochs
// (self.trainer.args.per_device_train_batch_size * self.trainer.args.gradient_accumulation_steps)
)
# Create new scheduler starting from current lr
# update the scheduler to accompany the new size
new_scheduler = get_cosine_schedule_with_warmup(
self.trainer.optimizer,
num_warmup_steps=0, # No warmup since we're continuing
num_training_steps=num_training_steps,
initial_lr=current_lr
)
self.trainer.lr_scheduler = new_scheduler
Margin Bandit Mining w/ Self Selection using FAISS:
We use the same bandit based structure from Margin Based Bandit Mining but instead of only selecting a random example, we use the same embedding strategy from Model-Based Mining to select the K most difficult hard-negatives using the model itself + any random noise we might want to add via random_n. So this one we basically have a combination of self reinforcement learning with faiss + reinforcement learning on the hard-negative parameters themselves. This was really cool and I think it’s a really good way to do bandit mining. The problem is it’s slow and incremental w/ respect to to choosing parameters. I think what I’ll probably do is take this method, and sort of figure out ‘scaling laws’ and see how to make good cold-start parameter choices given the known data/params/etc.
class AdaptiveBanditFaissCallback(TrainerCallback):
"""
1) Each epoch-end, re-embed entire doc_list using the current model => build a FAISS index
2) Maintain a local 'query_caches' for each query => do remove_easy + refill
using the FAISS index to find truly "hard" negatives
3) Re-build the trainer's dataset from caches => next epoch sees it
4) Use a bandit for dynamic 'remove_threshold'
"""
def __init__(
self,
trainer,
train_dataset,
model_path,
k_cache=5,
bandit_arms=None,
epsilon=0.1,
refill_every=2,
eval_metric_key="eval_loss",
epoch_start=1,
embedding_dim=768,
chunk_size=256,
create_hardnegs_fn=create_parallel_hard_negatives,
#create-hardnegs-function
original_dataset_path=None,
adaptive_dataset_path=None,
gpus_for_hardneg="6,7",
reload_after=True,
num_gpus_for_hardneg=2,
batch_size=256,
embedding_size=1536,
k_hard=1,
random_n=0,
):
super().__init__()
self.trainer = trainer
self.model_path = model_path
self.k_cache = k_cache
self.refill_every = refill_every
self.eval_metric_key = eval_metric_key
self.epoch_start = epoch_start
self.embedding_dim = embedding_dim
self.chunk_size = chunk_size
self.create_hardnegs_fn = create_hardnegs_fn
# dataset args
self.original_dataset_path = original_dataset_path
self.adaptive_dataset_path = adaptive_dataset_path
self.num_gpus_for_hardneg = num_gpus_for_hardneg
self.batch_size = batch_size
self.embedding_size = embedding_size
self.k_hard = k_hard
self.random_n = random_n
self.gpus_for_hardneg = gpus_for_hardneg
self.reload_after = reload_after
# 1) Bandit
if bandit_arms is None:
bandit_arms = [-0.1, -0.05, 0.0, 0.05, 0.1]
self.bandit = EpsilonGreedyBandit(arms=bandit_arms, epsilon=epsilon)
# 2) Basic data
# 2) Build initial caches, etc. (similar to your original code)
self.doc_list, self.doc_to_idx, self.idx_to_doc, self.query_idx_relations = create_document_list_with_relations(train_dataset)
self.query_caches = {}
for qid in self.query_idx_relations:
negs = list(self.query_idx_relations[qid]['negative']) # example
# pick up to k_cache
chosen = negs[:k_cache]
self.query_caches[qid] = [(nid, 999.0) for nid in chosen]
# For doc -> index row mapping:
# a simple approach: doc_id == index in doc_list
# Or if you have doc_to_idx, store that here.
self.epoch_counter = 0
self.remove_threshold = None
self.current_arm_index = None
# 4) Build initial training dataset from caches
self._build_train_dataset_from_caches()
def on_epoch_begin(self, args, state: TrainerState, control: TrainerControl, **kwargs):
self.epoch_counter += 1
if self.epoch_counter < self.epoch_start:
return
# Bandit picks threshold
self.current_arm_index = self.bandit.select_arm()
self.remove_threshold = self.bandit.get_arm_value(self.current_arm_index)
print(f"[BanditFaissCallback] Epoch={self.epoch_counter}, chosen threshold={self.remove_threshold:.3f}")
def on_epoch_end(self, args, state: TrainerState, control: TrainerControl, **kwargs):
"""
1) Re-embed entire doc_list => build FAISS index
2) remove_easy_batch
3) refill_caches_batch (using FAISS to get truly 'hard' negatives)
4) measure reward => update bandit
5) rebuild training dataset => next epoch
"""
if self.epoch_counter < self.epoch_start:
return
# (A) measure reward from log_history
reward = 0.0
for entry in reversed(state.log_history):
if self.eval_metric_key in entry:
reward = entry[self.eval_metric_key]
break
print(f"[BanditFaissCallback] Epoch={self.epoch_counter}, performance metric={reward:.4f}")
# (B) bandit update
self.bandit.update(self.current_arm_index, reward)
# (C) Only run on main process
if self._is_main_process(state):
# re-embed doc_list => build FAISS index
print(f"[BanditFaissCallback] Re-embedding entire doc_list to build FAISS index.")
self._build_faiss_index() # produce self.faiss_index, plus doc_id->faiss_id?
if dist.is_initialized():
dist.barrier()
# (D) remove easy negatives in batch (using the newly built index for margin computations)
self._update_caches_remove_easy_batch()
# (E) refill from FAISS (rather than random), picking truly "hard" negatives
if self.epoch_counter % self.refill_every == 0:
self._refill_caches_batch()
# (F) rebuild the trainer dataset
self._build_train_dataset_from_caches()
# ---------------------------------------------------------------
# FAISS index + doc embeddings
# ---------------------------------------------------------------
def _build_faiss_index(self):
"""
Re-encode all docs in doc_list, build a FAISS index in memory, store it as self.faiss_index.
We'll do something like:
embs = self.model.encode(doc_list, batch_size=..., convert_to_tensor=True)
self.faiss_index = build_faiss_index(embs, index_type="IndexFlatIP")
"""
# 3) Determine the current model checkpoint path
model_ckpt_path = self.model_path or self.trainer.args.output_dir
# we get the final checkpoint h
checkpoints = [d for d in os.listdir(model_ckpt_path) if d.startswith('checkpoint-')]
# Sort checkpoints by epoch number
checkpoints.sort(key=lambda x: int(x.split('-')[1]))
# get last checkpoint
checkpoint_path = os.path.join(model_ckpt_path, checkpoints[-1])
# 4) Call the user-provided function
self.create_hardnegs_fn(
dataset_path=self.original_dataset_path,
save_path=self.adaptive_dataset_path,
model_path=checkpoint_path,
num_gpus=self.num_gpus_for_hardneg,
batch_size=self.batch_size,
embedding_size=self.embedding_size,
k_hard=self.k_hard,
random_n=self.random_n,
)
self.faiss_index = faiss.read_index("embeddings.index")
self.faiss_to_doc_idx = pickle.load(open('faiss_to_doc_idx.pkl', 'rb'))
self.doc_id_to_faiss_id = {v: k for k, v in self.faiss_to_doc_idx.items()}
def _compute_margin_faiss(self, qid, neg_id, pos_id):
"""
We'll fetch embeddings from self.faiss_index by reconstruct(neg_id),
reconstruct(pos_id). For the query, we might store it or also do a dynamic approach.
For now, let's assume query is also in doc_list and qid is the same as doc_id.
If queries differ from doc_list, we might store them separately or do model.encode once.
This depends on how your queries are formed, but let's assume qid is a doc in doc_list.
"""
import numpy as np
# reconstruct doc embeddings from faiss
neg_vec = self.faiss_index.reconstruct(neg_id) # shape (d,)
pos_vec = self.faiss_index.reconstruct(pos_id)
q_vec = self.faiss_index.reconstruct(qid) # if queries are also among doc_list
# cos_sim
def cos_sim(a, b):
return np.dot(a, b) / (np.linalg.norm(a) * np.linalg.norm(b) + 1e-8)
cos_qn = cos_sim(q_vec, neg_vec)
cos_qp = cos_sim(q_vec, pos_vec)
margin = cos_qn - cos_qp
return margin
# ---------------------------------------------------------------
# Update + Refill
# ---------------------------------------------------------------
def _update_caches_remove_easy_batch(self):
"""
We do batch approach: gather all (qid, pos, neg) from caches => compute margin using FAISS
=> remove if margin < self.remove_threshold.
"""
self.faiss_index = faiss.read_index("embeddings.index")
self.faiss_to_doc_idx = pickle.load(open('faiss_to_doc_idx.pkl', 'rb'))
self.doc_id_to_faiss_id = {v: k for k, v in self.faiss_to_doc_idx.items()}
batch_q = []
batch_p = []
batch_n = []
index_map = []
for qid, rels in self.query_idx_relations.items():
pos_list = list(rels.get('positive', []))
if not pos_list:
continue
pos_id = pos_list[0] # assume 1 positive
cache_entries = self.query_caches[qid]
for idx, (neg_id, old_hardness) in enumerate(cache_entries):
batch_q.append(qid)
batch_p.append(pos_id)
batch_n.append(neg_id)
index_map.append((qid, idx))
if not batch_q:
return
# compute margins in a loop or in parallel
# We'll do it in a naive loop of size len(batch_q), though we could do partial reconstruct on GPU, etc.
margin_list = []
for (q, p, n) in zip(batch_q, batch_p, batch_n):
m = self._compute_margin_faiss(q, n, p)
margin_list.append(m)
# remove or keep
for i, (qid, cache_idx) in enumerate(index_map):
margin_val = margin_list[i]
if margin_val < self.remove_threshold:
self.query_caches[qid][cache_idx] = None
else:
self.query_caches[qid][cache_idx] = (self.query_caches[qid][cache_idx][0], margin_val)
# filter out removed
for qid in self.query_idx_relations:
self.query_caches[qid] = [x for x in self.query_caches[qid] if x is not None]
def _refill_caches_batch(self):
"""
Instead of random, we do a FAISS-based 'retrieve_hard_negatives' for each query that needs more negs.
"""
self.faiss_index = faiss.read_index("embeddings.index")
self.faiss_to_doc_idx = pickle.load(open('faiss_to_doc_idx.pkl', 'rb'))
self.doc_id_to_faiss_id = {v: k for k, v in self.faiss_to_doc_idx.items()}
needed_info = []
for qid, rels in self.query_idx_relations.items():
current_len = len(self.query_caches[qid])
needed = self.k_cache - current_len
if needed > 0:
needed_info.append((qid, needed))
if not needed_info:
return
# we can retrieve top-K from FAISS for each qid. Suppose queries are also in doc_list => same ID
for (qid, needed) in needed_info:
pos_list = list(self.query_idx_relations[qid].get('positive', []))
if not pos_list:
continue
pos_id = pos_list[0]
# retrieve top-K 'closest' docs to qid
cands = self._retrieve_hard_negatives_faiss(qid, top_k=needed*5) # retrieve some overshoot
# filter out qid itself, positives, or duplicates
filter_set = set(pos_list) | {qid}
cands = [d for d in cands if d not in filter_set]
cands = cands[:needed] # now we have 'needed' neg IDs
# compute margins + skip if margin < remove_threshold if you like
for doc_id in cands:
margin_val = self._compute_margin_faiss(qid, doc_id, pos_id)
if margin_val < self.remove_threshold:
continue
self.query_caches[qid].append((doc_id, margin_val))
def _retrieve_hard_negatives_faiss(self, qid, top_k=10):
"""
Simple example: we do a search in FAISS for the 'closest' docs to qid.
Because qid is also a doc in doc_list, we reconstruct qid embedding, do index.search(...).
"""
import numpy as np
q_vec = self.faiss_index.reconstruct(self.doc_id_to_faiss_id[qid])
# If the index is IndexFlatIP, we might want to L2-normalize q_vec or do it if the doc index is normalized
# We'll do a naive approach with index.search(1, q_vec)
# Actually index.search requires shape (n_query, dim), so we do q_vec as 1 row
query_batch = np.expand_dims(q_vec, axis=0)
distances, indices = self.faiss_index.search(query_batch, top_k + 50) # overshoot
# indices shape: (1, top_k+50)
# filter out qid if present
retrieved_ids = []
for faiss_idx in indices[0]:
doc_idx = self.faiss_to_doc_idx[faiss_idx]
if doc_idx != qid and doc_idx not in self.query_idx_relations[qid]['positive']:
retrieved_ids.append(doc_idx)
# done
return retrieved_ids
# ---------------------------------------------------------------
# Build dataset from caches
# ---------------------------------------------------------------
def _build_train_dataset_from_caches(self):
new_samples = []
for qid, rels in self.query_idx_relations.items():
pos_list = list(rels.get('positive', []))
if not pos_list:
continue
pos_id = pos_list[0]
q_text = self.doc_list[qid]
p_text = self.doc_list[pos_id]
for (neg_id, hardness) in self.query_caches[qid]:
neg_text = self.doc_list[neg_id]
new_samples.append({
"query": q_text,
"positive": p_text,
"negative": neg_text
})
ds = Dataset.from_list(new_samples)
self.trainer.train_dataset = ds
self.trainer._train_dataset = ds
self.trainer.train_dataloader = self.trainer.get_train_dataloader()
def _is_main_process(self, state: TrainerState):
is_main = True
fn = getattr(self.trainer, "is_world_process_zero", None)
if callable(fn):
is_main = self.trainer.is_world_process_zero()
elif hasattr(state, "is_world_process_zero"):
is_main = state.is_world_process_zero
return is_main
def build_new_training_lr_schedule(self, new_dataset):
current_lr = self.trainer.optimizer.param_groups[0]['lr']
# Calculate new total steps based on new dataset size
num_training_steps = (
len(new_dataset)
* self.trainer.args.num_train_epochs
// (self.trainer.args.per_device_train_batch_size * self.trainer.args.gradient_accumulation_steps)
)
# Create new scheduler starting from current lr
# update the scheduler to accompany the new size
new_scheduler = get_cosine_schedule_with_warmup(
self.trainer.optimizer,
num_warmup_steps=0, # No warmup since we're continuing
num_training_steps=num_training_steps,
initial_lr=current_lr
)
self.trainer.lr_scheduler = new_scheduler
Final Results
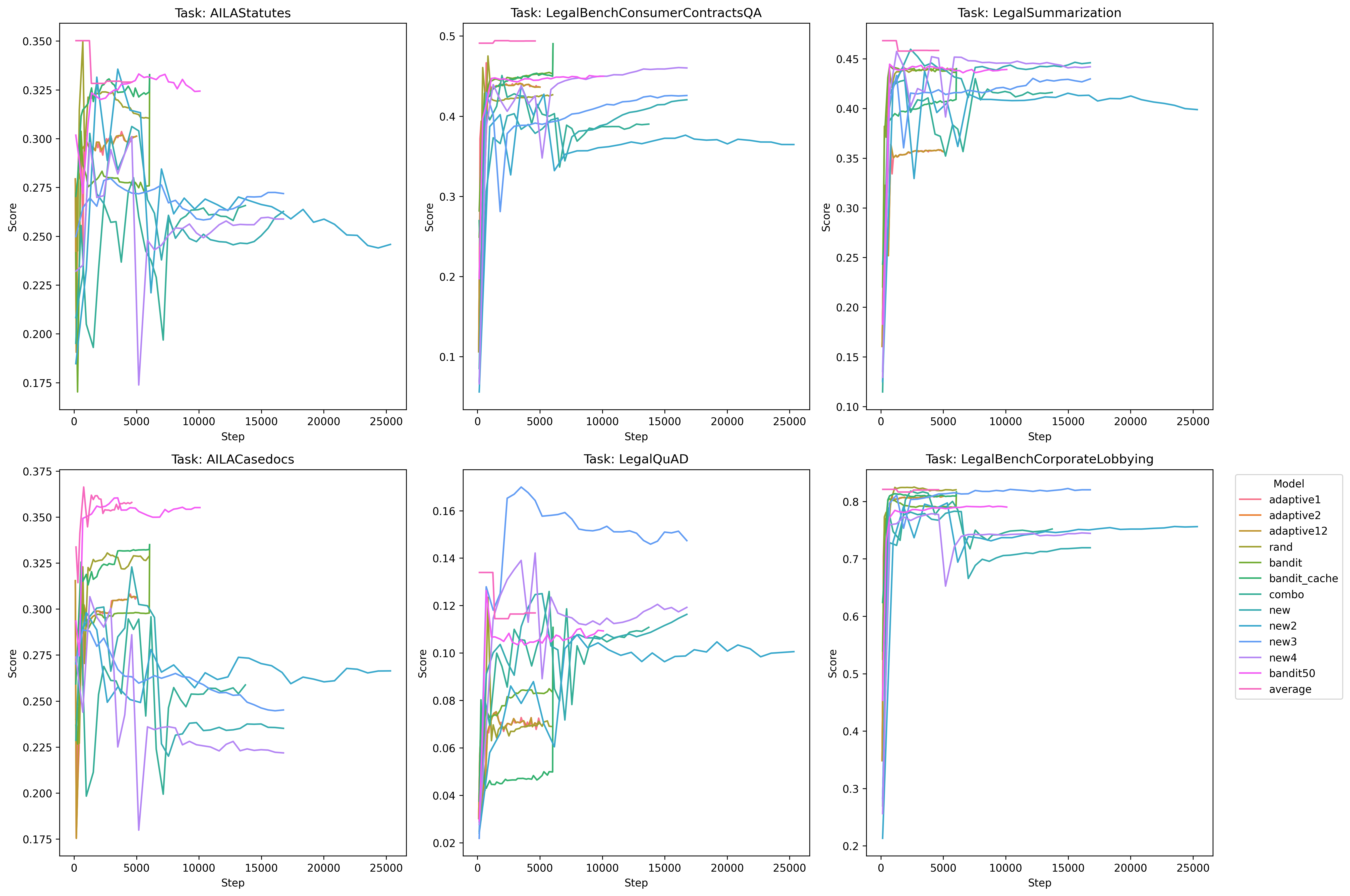
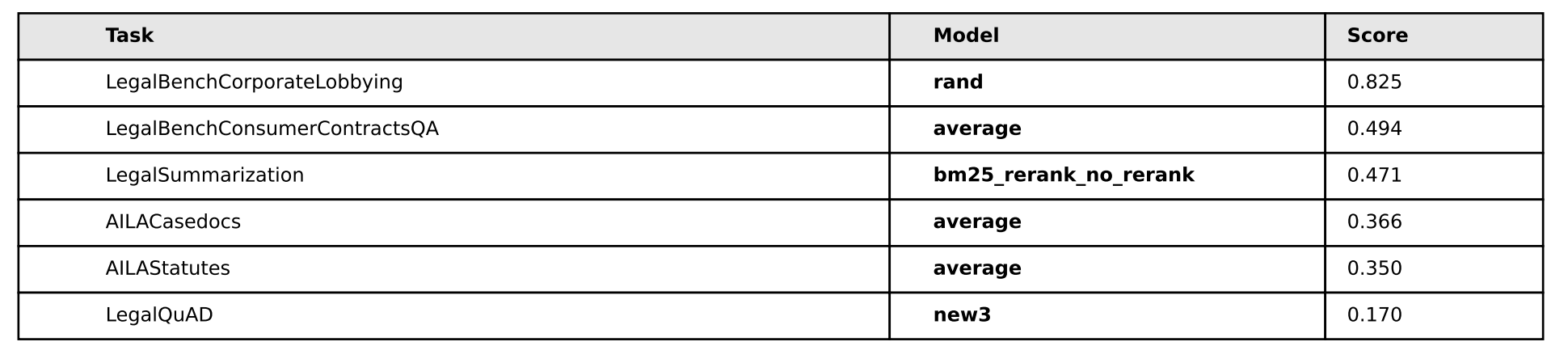
ok, so a few things to note:
- ‘rand’ is HardNegMiningCallback with k_hard=1 and 25 random samples with ‘start_epoch’ set to 10 (meaning we dont start mining until we’re at the 10th epoch). we train on the bm25 dataset for 10 epochs, then use the last-epoch’s model to find the most difficult negative and create a single triplet along with 25 other triplets where the hardnegative is chosen randomly. I wanted to add some less-difficult noise to our system. It produced pretty solid results and was usually the second performing model. I think the ultimate take away is probably the bm25 for 10 epochs more than anything.
- ‘new3’ is AdaptiveBanditFaissCallback with k_hard=3 and using the combo dataset of bm25 + rerank + no-rerank.
- the simple act of taking 3 models (bm25, no-rerank, rerank) and averaging their weights produces a very strong model; Im unclear of the mechaniics of why this is, but it’s pretty cool.
- for the task most similar to our training data (legal summarization), the cheapest option ‘bm25 + rerank + no-rerank’(eg, combine 3 datasets and have 3 examples per query/positive pair) produces the best model
- I didn’t get to explore the bandit models as much as I wanted to, but maybe I’ll write something a bit more after some of these other tasks finish training.
Admittedly this was pretty fun. We have a constrained dataset/problem that could honestly be solved with more data/compute (The Bitter Lesson, anyone?). But I wanted to pull various levers to measure effects and just sort of learn/find out what I can find out more about what could make this thing tick. And at some point you either do good science or you dont. So I think this is a good start.
TODO list (that I admitted don’t have a):
- Experiment with different embedding dimensions
- Compare performance across model sizes (small, base, large)
- Vary the similarity thresholds in hard negative selection
- Test different k values for nearest neighbor retrieval
- Compare performance with and without embedding normalization
- Analyze performance with different batch sizes
- Study the impact of learning rates
- Investigate gradient accumulation steps
- Test different loss functions (InfoNCE, triplet loss variants)
- Combine BM25 and reranking with different weighting schemes
- Test ensemble methods of multiple rerankers
- Explore sparse-dense retrieval combinations